Abstract
Arsenic and antimony are metalloids with profound effects on biological systems and human health. Both elements are toxic to cells and organisms, and exposure is associated with several pathological conditions including cancer and neurodegenerative disorders. At the same time, arsenic- and antimony-containing compounds are used in the treatment of multiple diseases. Although these metalloids can both cause and cure disease, their modes of molecular action are incompletely understood. The past decades have seen major advances in our understanding of arsenic and antimony toxicity, emphasizing genotoxicity and proteotoxicity as key contributors to pathogenesis. In this review, we highlight mechanisms by which arsenic and antimony cause toxicity, focusing on their genotoxic and proteotoxic effects. The mechanisms used by cells to maintain proteostasis during metalloid exposure are also described. Furthermore, we address how metalloid-induced proteotoxicity may promote neurodegenerative disease and how genotoxicity and proteotoxicity may be interrelated and together contribute to proteinopathies. A deeper understanding of cellular toxicity and response mechanisms and their links to pathogenesis may promote the development of strategies for both disease prevention and treatment.
Similar content being viewed by others
Introduction: chemistry and biology of arsenic and antimony
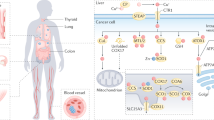
Arsenic and antimony are classified as metalloids, that is, chemical elements that possess physical and chemical properties that are intermediate between metals and nonmetals. These metalloids are present in trace amounts in the Earth’s crust (mean concentrations: 1–2 ppm for arsenic (13–26 µM) and 0.2 ppm for antimony (1.6 µM)) but can reach high local concentrations due to geological sources or anthropogenic activities [1, 2]. For example, arsenic concentrations in the groundwater from geological sources reaching up to or above 5000 ppb (67 µM) have been measured at sites in Bangladesh, China, Thailand, Argentina, and Australia [3,4,5], greatly exceeding the levels in drinking water (10 ppb; 0.13 µM) recommended by the World Health Organization (WHO). Similarly, high antimony concentrations have been measured in water and sediments close to mining and smelting areas in China (up to 29,000 ppb; 240 µM), raising concerns that antimony concentrations in the groundwater in these areas may exceed the recommended level in drinking water recommended by WHO (10 ppb; 0.08 µM) [2, 6, 7]
The presence of arsenic in soils and water is a major human health threat, and is estimated to affect up to 200 million people worldwide [8]. Arsenic can accumulate and damage every organ in the human body with the highest concentrations present in the liver and kidney [9]. Chronic arsenic exposure is associated with several pathological conditions including skin disorders, cardiovascular disease, diabetes, cancers, as well as neurological and neurodegenerative disorders [3, 10,11,12,13]. Antimony has also been proposed to be a global health threat [7]. Chronic antimony exposure may affect the skin, the respiratory, cardiovascular and gastrointestinal systems, and possibly cause cancer [14, 15]. The highest concentration of antimony in humans is found in liver and kidney, and certain inhaled antimony compounds are retained in lung for long periods [7, 16].
Arsenic concentrations reported in epidemiological studies to cause adverse health effects usually range from 100 to 1000 ppb (1.3–13 µM) and some studies suggest that it may already affect human health below 10 ppb (0.13 µM) [9, 17]. The majority of the cellular studies cited in this review exposed various human cell lines to concentrations ranging from 10 to 100 µM (750–7500 ppb), corresponding to exposure to high environmental arsenic concentrations. Several cellular studies have also used lower arsenic concentrations (from 30 nM up to 10 µM; 5–750 ppb) that correspond to low to moderate environmental arsenic. As a comparison, the median arsenic concentration in healthy humans is in the range of 30–40 ppb (0.4–0.5 µM) in kidney and liver, 50–90 ppb (0.67–1.2 µM) in lung and skin, 10–30 ppb (0.13–0.4 µM) in brain, and 300–900 ppb (4–12 µM) in hair and nail [9]. Tissues taken from arsenic-intoxicated humans show much higher arsenic concentrations: 147,000 ppb (1.9 mM) in liver, 27,000 ppb (360 µM) in kidney, and 11,000 ppb (146 µM) in lung and brain [9]. Mean arsenic concentrations in the plasma and placenta of chronically exposed human populations are about 10 and 100 nM, respectively, while urinary concentrations of exposed individuals can reach up to 5650 ppb (75 µM) [18,19,20,21]. Finally, arsenic concentrations in the plasma of individuals treated with arsenic trioxide (As2O3) peaks at 5–7 µM (400–600 ppb) [22]. Thus, most toxicity mechanisms, both at epidemiological and cellular level, have been studied at moderate to high arsenic concentrations (µM range) while less is known about toxicity targets and mechanisms at low (nM) concentrations.
Epidemiological studies on the health effects of antimony in environmentally relevant concentrations are scarce and mainly concern occupational exposure in industrial settings and during treatment with antimony-containing drugs. The antimony concentration in the lungs of smelter workers exposed to antimony is about 2.6 μM, while the concentration of unexposed individuals is 0.2 μM [16]. The concentration of the pentavalent antimonial drug Glucantime® used to treat leishmaniasis is usually 20 mg/kg body weight per day, and plasma concentrations of antimony peak at 1 mM after injection of the drug [23]. The studies cited in this review usually exposed human cell lines to antimony concentrations ranging from 50 to 800 µM (750–7500 ppb), which corresponds to high exposure levels. Only few studies have explored toxicity mechanisms at low antimony concentrations.
Despite their toxicities, arsenic- and antimony-based drugs have been used for centuries in the treatment of multiple diseases, and current use includes the treatment of acute promyelocytic leukemia (APL) and of diseases caused by certain protozoan parasites; however, the molecular mechanisms of action of arsenic- and antimony-based drugs are in many cases still unclear [10, 24,25,26]. In spite of its in vitro anticancer activity, arsenic has not been shown to be effective in treating cancer types other than APL in clinical trials [27]. However, its efficacy in anticancer therapy may potentially be increased when combined with other anticancer drugs [28]. Several antimony-based compounds also show marked in vitro antiproliferative activity against a variety of human cancer cells [29, 30]. While antimony-based compounds have not yet been evaluated as anticancer drugs in clinical trials, their lower toxicity compared to arsenic-based compounds illustrates their future potential.
The molecular mechanisms of arsenic and antimony toxicity are only partially understood. In general, toxicity of a metal is governed by its physicochemical properties and ligand preferences. Arsenic and antimony are ‘hard’ transition metals that preferentially interact with oxygen in their higher oxidation states and sulfur in their lower oxidation states. The most common oxidation states are pentavalent and trivalent arsenic and antimony, with the trivalent states being more toxic than the pentavalent states [3, 7, 9, 16]. Typically, metals cause toxicity in vivo by interfering with protein and membrane functions, with nutrient uptake and redox reactions, and by causing DNA damage [31, 32]. At the cellular level, arsenic toxicity has been attributed to oxidative stress, changes to the epigenome, genotoxicity, and to altered protein function and activity. At the molecular level, pentavalent arsenate [As(V)], being chemically similar to phosphate, can cause toxicity by competing with phosphate in various biochemical reactions and by impairing energy generation via disruption of adenosine triphosphate (ATP) production. The toxicity of trivalent arsenite [As(III)] is mainly attributed to its high affinity for sulfhydryl groups, and binding of As(III) to reduced cysteine residues in proteins may disturb protein conformation, function, and interactions [32,33,34,35,36]. The ability of As(III) to bind to proteins is not only central for its toxic mode of action but can also contribute to its therapeutic effects. For example, binding to cysteine residues in the oncoprotein PML-RARα underlies the anticancer activity of arsenic trioxide in patients with APL [37, 38]. Similarly, As(III) binding to specific kinases and transcriptional regulators is associated with enhanced arsenic resistance in yeast and bacteria [39,40,41].
Compared to arsenic, much less is known about the mechanisms of antimony toxicity. It is generally assumed that arsenic and antimony are structurally related, and that they therefore act in similar ways. Like As(III), antimonite [Sb(III)] has been proposed to affect the cellular redox balance, to disrupt enzyme function by binding to sulfhydryl groups in proteins, and to cause DNA damage [15, 42,43,44]. Additionally, Sb(III) binding to proteins is not only associated with toxicity but also with antimony resistance in yeast and bacteria [39, 45, 46].
Metalloid toxicity is also dependent on metabolic transformations. Arsenic and antimony can undergo biomethylation in many microorganisms and mammalian species, including humans. However, the extent of biomethylation varies between species and may also give rise to species-specific compounds [44, 47, 48]. In humans, arsenic methylation primarily occurs in the liver and involves the arsenic (+ 3 oxidation state) methyltransferase enzyme As3mt [48]. Whether antimony is methylated in humans is unclear [44]. Nevertheless, antimony can be methylated in vitro by the human As3mt orthologue ArsM, isolated from the red alga Cyanidioschyzon merolaeis [49], raising the possibility that methylation might occur also in humans. Methylation changes the properties and behavior of arsenic and antimony and affects their modes of action and toxicities with methylated forms considered to be more toxic than the inorganic forms [48]. One way methylation affects metalloids is by modulating their binding to proteins: inorganic As(III) can bind up to three cysteine residues, monomethylarsonous acid [MMAs(III)] can bind two cysteine residues and dimethylarsonous acid [DMAs(III)] can bind only one cysteine residue [33]. Studies in budding yeast (Saccharomyces cerevisiae), that possesses a methyltransferase enzyme involved in As(III) methylation [50], indicated that As(III) methylation also serves a signaling function in an adaptative response to arsenic stress [50, 51]. Thus, depending on the form of the metalloid and on the specific target, arsenic and antimony binding to proteins may cause toxicity or resistance.
Studies in the past decades have greatly expanded our mechanistic understanding of arsenic and antimony toxicity and emphasized genotoxicity and proteotoxicity as key contributors to pathogenesis. Our understanding of how cells resist and adapt to metalloid toxicity has also increased substantially in recent years. In this review, we highlight mechanisms by which arsenic and antimony cause genotoxicity and proteotoxicity as well as mechanisms used by cells to maintain proteostasis during metalloid exposure. We also address how metalloid-induced proteotoxicity may promote neurodegenerative disease and how genotoxicity and proteotoxicity may be interrelated and contribute to proteinopathies in a concerted way.
How arsenic and antimony cause genotoxicity
Over the past decades, a large number of epidemiological studies documented a strong association between environmental, occupational, and medical exposure to arsenic and cancers of the skin, lung, and urinary bladder [52, 53]. Arsenic exposure has also been linked to the development of kidney, liver, and prostate cancer [53,54,55,56]. Consequently, arsenic, and its inorganic compounds, is classified as a group 1 human carcinogen [53]. Paradoxically, arsenic exhibits a weak mutagenic potential, even at toxic concentrations, and its carcinogenic properties failed to be reproduced in experimental animals for many years, obscuring its mechanisms of carcinogenesis [57, 58]. More recent studies, using transplacental/early-life or whole-life exposure approaches, found that inorganic arsenic has the ability to induce several types of cancer in mice [58,59,60]. Additionally, arsenic is a well-established co-carcinogen that potentiates the carcinogenic effects of ultraviolet radiation and benzo[a]pyrene present in tobacco smoke, and enhances the mutagenicity and clastogenicity of known genotoxins, such as cisplatin, methyl methanesulfonate, N-methyl-N-nitrosourea, and phleomycin [17, 61, 62]. Several, mutually non-exclusive, mechanisms of arsenic-elicited carcinogenesis have been proposed, reflecting arsenic’s multifaceted effects at the cellular level: these include (a) induction of oxidative stress resulting in increased oxidative DNA damage; (b) biotransformation of inorganic arsenic to methylated forms with higher genotoxicity; (c) inhibition of DNA repair mechanisms via transcriptional repression of genes encoding DNA repair enzymes and by modulating their enzymatic activity; (d) non-genotoxic epigenetic dysregulation (DNA methylation, post-translational histone modifications, miRNA) and aberrant activation of signal transduction pathways, which lead to expression changes of tumor suppressor genes and proto-oncogenes at the level of transcription initiation and alternative splicing, ultimately resulting in stimulation of cell proliferation and inhibition of apoptosis [17, 52, 63, 64]. Moreover, studies in budding yeast indicated that arsenic can generate DNA damage independently from oxidative stress, suggesting that additional modes of arsenic genotoxicity exist [62].
The genotoxic and carcinogenic potential of antimony is much less studied. The few available epidemiological studies of occupational and environmental exposure to antimony found either no association with cancer incidence or an increased risk of lung cancer [44, 65]. However, a rodent inhalation study revealed that Sb(III), in form of antimony trioxide, has the potential to induce lung and adrenal gland tumors as well as lymphomas [66]. Consequently, Sb(III) is currently categorized as probably carcinogenic to humans (group 2A) [67]. Like for arsenic, the carcinogenic properties of Sb(III) may result from induction of oxidative stress, inhibition of DNA repair mechanisms and changes in gene expression, leading to increased oxidative DNA damage and stimulation of cell proliferation [44]. Additionally, studies in S. cerevisiae revealed that Sb(III) also disrupts telomere maintenance and causes topoisomerase I (Top1)-dependent replication-associated DNA damage [68].
Up to now, the molecular mechanisms of metalloid-induced DNA damage and carcinogenesis remain elusive and still under debate. This section will highlight how As(III) and Sb(III) cause genotoxicity by partly distinct mechanisms.
Arsenic and antimony-induced genotoxic effects in mammalian cells
As(III) is inactive or extremely weak in its ability to induce gene mutations [63]. Moreover, there is no solid evidence for the reactivity of As(III) with DNA. However, it is well known that in cultured human cells, long-term exposure to As(III) at moderate to high concentrations (≥ 5 μM for 16–24 h) causes oxidative modifications of DNA and oxidative damage-related lesions, such as 8-hydroxyl-2′-deoxyguanine (8-OHdG), DNA breaks and DNA–protein crosslinks, as well as sister chromatid exchanges and chromosomal aberrations, including micronuclei and aneuploidy [52, 63]. Accordingly, several studies showed that As(III) increases production of reactive oxygen species (ROS) and reactive nitrogen species (RNS), whereas As(III)-induced oxidative DNA damage can be attenuated by the addition of antioxidants [69, 70]. Elevated levels of urinary 8-OHdG in individuals chronically exposed to high As(III) concentrations further supports the notion that high doses of As(III) cause DNA damage by increasing oxidative stress [71] (Fig. 1). Interestingly, induction of ROS production and increased oxidative DNA damage was also observed in the breast cancer cell line MCF-7 exposed to non-cytotoxic concentrations of As(III) (2 μM for 4 h) [72]. However, identical conditions did not increase ROS production in mouse embryonic fibroblasts (3T3) and three human cell lines (HeLa, HEK 293, HEMn-LP), providing evidence against the oxidative stress theory of arsenic carcinogenesis during chronic exposure to low levels [73].
How arsenic and antimony cause genotoxicity. Both in yeast and mammalian cells, As(III) and Sb(III) induce oxidative stress to varying degrees. This results in elevated levels of oxidative DNA damage, including oxidized bases and single-strand breaks (SSBs), which can also arise indirectly from incomplete repair of oxidized bases by base excision repair. SSBs can be converted to double strand breaks (DSBs) during replication or when SSBs are closely spaced. In addition, As(III) and Sb(III) increase the formation of protein–DNA adducts such as topoisomerase 1 (TOP1) DNA–protein crosslinks, either in a DNA oxidative damage-dependent manner or by interfering with TOP1 enzymatic activity. The presence of DNA–protein crosslinks leads to generation of replication-associated DSBs and single-stranded DNA (ssDNA) gaps. In budding yeast, oxidative stress and replication-independent DNA damage were also observed after metalloid treatment. In addition, As(III) and Sb(III) not only induce DSBs and ssDNA gaps, but also inhibit repair of these lesions by interfering with DNA damage repair pathways such as homologous recombination (HR), non-homologous end joining (NHEJ) and DNA damage tolerance (DDT). As(III) and Sb(III)-mediated disruption of the actin and microtubule cytoskeleton may also interfere with various aspects of DNA damage repair and cause chromosome aberrations. Finally, both metalloids perturb homeostasis of telomeres by oxidation of guanine-rich telomeric repeats, possibly by directly binding to telomeric DNA or by interfering with the function of telomere-associated proteins. This leads to telomere uncapping resulting in telomere erosion and fusion of chromosome ends. The figure was created with BioRender.com
Oxidation of bases in DNA and single-strand breaks (SSBs) are formed directly from hydroxyl radical attack. Additionally, SSBs can be generated during the repair of oxidized bases by base excision repair (BER). Furthermore, replication of DNA containing unrepaired SSBs results in the formation of double-strand breaks (DSBs) leading to chromosomal aberrations and genomic instability. Indeed, As(III) at concentrations of 5–50 μM induced replication-dependent DSBs in cultured AA8 Chinese hamster ovary cells, which activated the DNA damage response (DDR) followed by homologous recombination (HR) repair [74]. Moreover, in HR-deficient cells, DSBs were detectable already at 1 μM As(III) [74]. Additionally, closely spaced SSBs can also generate DSBs in the absence of replication, but this requires very high levels of oxidative damage and/or region-specific accumulation of such lesions. How As(III) generates oxidative stress-dependent DNA damage is not well understood. Metalloids are not redox-active elements, like iron (Fe) or copper (Cu), so it seems that they must induce ROS production indirectly. Early studies showed that ROS generated in As(III)-treated cells are mainly produced by membrane-bound NADPH oxidase (NOX), due to expression upregulation of NOX complex components [75, 76]. Interestingly, depletion of NOX components in leukemic cells led to nearly complete inhibition of ROS production by As(III) [76]. Other studies showed that As(III) at the concentrations of 15–10 µM also targets mitochondria, leading to the release of the superoxide anion and its subsequent reaction with nitric oxide to produce the less reactive and more stable peroxynitrite, which has the capacity to act throughout the cell [77, 78]. 2 μM As(III) was also able to increase the levels of nitric oxide by upregulating the expression of nitric oxide synthase (NOS) in human keratinocytes [79]. Other types of ROS generated by As(III) via NOX or mitochondria, such as H2O2, are too reactive and unstable to penetrate the nucleus and induce oxidative DNA damage [80]. Methylated forms of arsenic are a potential source of ROS that can be generated within the nucleus. It has been demonstrated that the reaction between molecular oxygen and DMAs(III) produces dimethylarsenic peroxyl radical and superoxide anion radical [81]. Importantly, methylation of As(III) and subsequent generation of dimethylarsenic peroxyl radical likely contributes to induction of DNA breaks in lung cells of rodents and human alveolar epithelial type II (L-132) cells [82]. Finally, in human CD4 T lymphocytes, As(III) at the cytotoxic concentration of 5 μM has recently been shown to induce the accumulation of topoisomerase 1-DNA covalent cleavage complexes (TOP1cc) [83], which, in turn, can stall replication fork progression leading to generation of DSBs. Importantly, TOP1cc can be trapped on DNA due to inhibition of TOP1 activity or by various DNA lesions, including oxidative DNA damage [84].
Another source of DNA damage, such as chromosomal aberrations and aneuploidy in cells exposed to As(III), may be dysfunctional telomeres (Fig. 1). Epidemiological studies reported both lengthening and shortening of telomeres in individuals chronically exposed to As(III) [85]. A possible mechanism for As(III)-induced telomere lengthening involves the upregulation of the telomerase catalytic subunit (hTERT) and telomere binding proteins such as TRF1 and TRF2, subunits of the telomere sheltering complex [85]. Thus, As(III) may partially contribute to carcinogenesis by promoting telomere elongation, which is crucial for the maintenance and viability of human cancer cells. Paradoxically, As(III)-induced downregulation of both hTERT and TRF2 was also observed and associated with telomere shortening. Since telomere sequences are rich in guanine, which is particularly sensitive to oxidative damage, it has been proposed that As(III) may cause telomere attrition through oxidative stress [85]. Importantly, 4 μM As(III) induced increased ROS levels in glioblastoma cells (U87 line) followed by translocation of telomerase from the nucleus to the cytoplasm, telomere shortening, and accumulation of γH2AX (histone H2AX variant phosphorylated on Ser139, which serves as a sensitive marker of DNA damage and repair) that co-localized with TRF1, strongly suggesting that the telomeres are sites of As(III)-generated DNA damage in an oxidative stress-dependent manner [86]. Activation of DDR at telomeres and erosion of telomeres accompanied by downregulation of TRF2 expression were also observed in human CD4 T lymphocytes exposed to 5 μM As(III) [83]. A study in CD1 mouse embryos revealed that 30 μM As(III) induced telomere erosion and end-to-end chromosome fusions, which was prevented by co-administration of an antioxidant [87]. Interestingly, telomerase-deficient mouse embryos with shortened telomeres exhibited decreased resistance to As(III) [87]. Moreover, prostate cancer cells (PC-3 line) characterized by very short telomeres were about 10- to 100-fold more sensitive to As(III) compared to human cell lines with longer telomeres [88]. Another study also reported an association between longer telomere lengths and increased resistance to As(III) in human cells in vitro [89]. In PC-3 prostate cancer cells, cytotoxic concentrations of As(III) (0.23–0.45 μM) triggered telomere dysfunction, manifested by telomere-associated accumulation of γH2AX and telomere degradation, possibly by directly binding to telomeric TTAGGG repeats, resulting in displacement of hTERT from telomeres [88]. Importantly, telomeric DNA damage was observed in the absence of oxidative DNA damage in PC-3 cells, suggesting that oxidative stress-independent telomere dysfunction may be a key source of As(III)-induced genotoxicity in cells with short telomere length [88].
Unlike for As(III), there are limited data on the genotoxic potential of Sb(III). Sb(III) did not induce mutations either in bacterial assays or in cultured mammalian cells [44], although one early report showed a positive response to SbCl3 in the Bacillus subtilis Rec-assay [90]. Many studies in mammalian cell cultures or rodents have shown no clastogenic effects of Sb(III) [44, 68]. Moreover, 50 μM SbCl3 did not induce any detectable DSBs in HeLa S3 cells after 8 h treatment [91]. On the other hand, exposure of HepG2 and LS-174 T human cancer cell lines to high concentrations of SbCl3 (100–500 μM) resulted in the induction of the DNA damage marker γH2AX [92]. Some historical studies reported clastogenic activity of Sb(III), such as induction of micronuclei, sister chromatid exchange and chromosomal aberrations [44, 68]; however, these studies have recently been inspected according to the applicable standards and have turned out to be mostly inconclusive and of poor quality [44]. A more recent analysis of the genotoxic properties of antimony compounds by the ToxTracker assay revealed that low concentrations of Sb(III) (0.2–0.5 μg/mL or 0.3–0.8 μM) have the potential to induce oxidative stress-derived DNA damage but has no ability to directly damage DNA or interfere with replication [93]. 1 μg/mL (8 µM) of Sb(III) also activated the unfolded protein response suggesting induction of protein stress [93]. Very high concentrations of Sb(III) (105 μg/mL; 860 μM) strongly disrupted redox homeostasis in human THP-1 macrophages, leading to a 50% decrease in intracellular free glutathione (GSH) levels, due to the formation of Sb(GS)3 conjugates as a mean of Sb(III) detoxification, and due to Sb(III)-mediated inhibition of glutathione reductase catalyzing the regeneration of GSH from oxidized GSSG [94]. SbCl3 at the concentration of 60 μM was demonstrated to cause death of rat hepatocytes by generating ROS [95]. Furthermore, in zebrafish liver, high concentrations of Sb(III) (16.58 and 33.16 mg/L) induced oxidative stress resulting in the accumulation of oxidative DNA damage but not DNA–protein cross-links [96]. In summary, Sb(III) appears to exhibit an indirect genotoxic mode of action involving increased oxidative stress at relatively high concentrations, but further research is needed to firmly establish Sb(III) as a genotoxin for animals, including humans.
Studies of arsenic and antimony genotoxicity in budding yeast
S. cerevisiae is an excellent model organism to study various aspects of DNA damage response, DNA repair pathways, and mechanisms of DNA damage induction by chemical agents [97,98,99]. Studies using either wild-type yeast or mutants devoid of As(III)/Sb(III) detoxification transporters Acr3 and Ycf1 revealed that both As(III) and Sb(III) exhibit genotoxic properties at subcytotoxic concentrations [62, 68]. 1 h exposure to As(III) triggered phosphorylation of histone H2A on Ser129 (an equivalent of human γH2AX, which is a sensitive DNA damage marker also in yeast) [97,98,99], starting from 250 μM in wild type and 50 μM in the double mutant acr3Δ ycf1Δ [62, 68]. For Sb(III), high levels of H2A phosphorylation were observed after 1–2 h treatment with 200–500 μM in ycf1Δ cells [62, 68]. In contrast to mammalian cells, subcytotoxic or cytotoxic concentrations of both As(III) (0.5–1 mM) and Sb(III) (0.2–5 mM) induced low levels of ROS and only a small increase in oxidative DNA damage [62, 68]. Moreover, a BER-defective mutant showed a very mild sensitivity to either metalloid, strongly suggesting that As(III) and Sb(III) have weak potential to generate oxidative stress-dependent DNA damage in budding yeast. Also, cells defective in nucleotide excision repair (NER) showed wild type resistance to both metalloids, which indicates that As(III) and Sb(III) do not generate bulky DNA adducts in yeast. In contrast, HR- and DNA damage tolerance (DDT)-defective yeast mutants devoid of DSB and replication-associated damage repair, respectively, were substantially As(III) and Sb(III) sensitive. Consequently, As(III) and Sb(III)-treated S phase cells showed increased accumulation of SSBs and ssDNA gaps, as well as Rad52 DNA repair nuclear foci, which indicate ongoing DNA repair by HR [62, 68] (Fig. 1). For Sb(III), the levels of SSBs, ssDNA regions and Rad52 foci were partially reduced in cells lacking Top1, but not by the addition of a ROS scavenger [68]. This indicates that a subset of Sb(III)-induced replication lesions may be generated due to the formation of Top1-DNA adducts, but independently from oxidative stress (Fig. 1). Interestingly, induction of Top1-induced DNA fragmentation by As2O3 (1–2 μM for 72 h) has also been reported in human leukemia NB4 cells, but in an oxidative stress-dependent manner and accompanied by apoptosis activation [100].
Sensing of DSBs or ssDNA gaps by the upstream kinases of the DDR, Mec1 and Tel1, results in phosphorylation of histone H2A and hyperphosphorylation of the effector kinase Rad53, which serve as sensitive markers for these types of DNA damage [97]. Importantly, both As(III) and Sb(III) induce high levels of H2A and Rad53 phosphorylation, not only in S phase but also in G2/M phase, suggesting DSB induction in the absence of replication and elevated oxidative stress (Fig. 1). In G1 phase-synchronized cells devoid of the Ku complex, that binds to and protects DSBs from resection and thus prevents activation of DDR in G1, H2A and Rad53 phosphorylation was also observed after As(III) and Sb(III) treatment. However, the presence of DSBs was detectable only at a very high concentration of As(III) (25 mM) [62, 68]. Thus, it remains to be established what types of DNA lesions are mainly responsible for As(III)-induced activation of DDR in yeast at subcytotoxic concentrations. Moreover, no presence of DSBs was detected upon Sb(III) treatment of yeast cells [68]. Instead, Sb(III) negatively affects telomere stability, since mutants devoid of proteins involved in telomere maintenance, such as Cdc13, Tel1, and Yku70, were found to be highly sensitive to Sb(III) [68]. Importantly, Sb(III) sensitivity of the cdc13-1 mutant was strongly supressed by deletion of the PIF1 or EXO1 genes encoding nucleases contributing to resection of uncapped telomeres. The cdc13-1 mutant also showed high sensitivity to As(III), which suggests that both metalloids are able to disrupt homeostasis of telomeres leading to DDR activation [68] (Fig. 1). The fact that metalloid-induced DNA damage in yeast cells is largely independent of oxidative stress and also observed in the absence of replication, suggests that both As(III) and Sb(III) might also exhibit in situ DNA damage activity. We hypothesize that both metalloids might generate low levels of sequence- or chromosome site-specific DSBs, which are not detectable by standard techniques.
However, the question remains whether the mechanisms of As(III) and Sb(III) genotoxicity observed in budding yeast may be similar in human cells. Compared to mammals, yeast cells are more resistant to both metalloids, in part due to the presence of efficient detoxification pathways mediated by the plasma membrane As(III)/Sb(III) efflux transporter Acr3 and the ATP-binding cassette (ABC) transporter Ycf1, which sequesters As(III)/Sb(III)-glutathione conjugates into the vacuole [101]. Indeed, the use of yeast mutants lacking these transporters allowed to decrease concentrations of As(III) and Sb(III) exerting genotoxic effects by five- and tenfold, respectively [62, 68]. Members of the Acr3 family are absent in animals; however, mammalian cells express a wide plethora of plasma membrane-localized ABC transporters, which pump free or glutathione-conjugated arsenic species, including inorganic As(III), MMAs(III) and dimethylarsinic acid [DMAs(V)] out of the cells, enabling arsenic clearance from the whole body [101]. Upregulation of these transporters is the key mechanism of acquired high-level As(III) resistance observed in cancer cells [101]. Both yeast and mammals can also limit intracellular accumulation of As(III) and Sb(III) by downregulating expression or activity of aquaglyceroporins and sugar transporters, which serve as entry pathways for As(III) and Sb(III) in all kingdoms of life [101]. On the other hand, these channels and transporters were also shown to extrude methylated forms of arsenic in mammalian cells. Interestingly, it has been recently demonstrated that budding yeast also produce MMAs(III) and DMAs(III), which contribute to the toxic effects of inorganic As(III) exposure [50, 51]. Whether the production of methylated As(III) species increases DNA damage in yeast cells remains to be investigated. Higher resistance of yeast cells to As(III) and Sb(III) compared to human cells may also be explained by the fact that budding yeast lacks certain DNA repair proteins and cycle regulators, such as the poly(ADP-ribose) polymerase-1 (PARP-1) protein, BRCA1 and p53, which are known to be targeted by these metalloids (see below). On the other hand, Chinese hamster ovary (CHO-K1) cells, often used in metalloid genotoxicity studies, are about 10 times more resistant to As(III) than human fibroblasts [102]. Even human cell lines can vary greatly in susceptibility to As(III) [88]. Differences in arsenic resistance between animal species may be due to the length of telomeres [87], antioxidant capacity [102] and dissimilar rates and patterns of arsenic methylation [103, 104]. Nevertheless, it should be emphasized that DNA damage caused by As(III) and Sb(III) in all model organisms and cell lines is observed mainly at species-appropriate subcytotoxic and cytotoxic concentrations and it remains to be determined whether or how the genotoxic potential of As(III) and Sb(III) contributes to the development of cancer in humans during chronic exposure to low levels of metalloids.
Inhibition of DNA repair by arsenic and antimony
It is well documented that low concentrations of As(III) potentiates DNA damage induced by several physical and chemical agents in mammalian cells via indirect or direct inhibition of DNA repair pathways, and thus may contribute to genomic instability and, ultimately, cancer (Fig. 1). A number of studies reported that As(III) treatment results in decreased expression of several DNA repair genes involved in BER and NER, as observed in mammalian cell lines and in cells isolated from individuals chronically exposed to As(III) [17, 105]. As(III)-induced downregulation of gene expression likely involves changes in DNA methylation and histone modification patterns. For example, 1 μM As(III) reduces expression of the DSB repair BRCA1 gene by inducing hypermethylation of its promoter in MCF7 breast cancer cells [106], while dose-dependent decrease in expression of several BER genes (MPG, PARP1, XRCC1) in human keratinocytes (HaCaT cells) treated with subcytotoxic and cytotoxic concentrations of As(III) (2.5–10 μM) was associated with the H3K9me2 heterochromatin mark enrichment in their promoter regions [107]. Decreased expression of some mismatch repair (MMR) genes, such as MLH1 and MSH2, due to promoter DNA hypermethylation was observed in HaCaT cells treated with 0.1–0.5 μM As(III) [108] and in peripheral blood mononuclear cells isolated from humans chronically exposed to As(III) in drinking water [109].
As(III) can also directly impact the activity of DNA repair proteins. As(III) has been shown to bind to zinc finger (ZnF) motifs, which are common in DNA-binding proteins, including transcription factors and DNA repair proteins, as well as in so-called Really Interesting New Gene (RING) domain proteins such as E3 ubiquitin ligases [110]. Mass spectrometry analysis revealed that As(III) selectively displaces Zn(II) from C3H1 and C4 ZnF motifs, but not from the more common C2H2 motif, and that As(III) is coordinated by three cysteine residues [111]. In contrast, MMAs(III) binds to two cysteines and can thus interact with all types of ZnF motifs [112]. Arsenic-bound ZnF motif-containing peptides display an altered conformation, suggesting that As(III) binding may affect the function of ZnF proteins [113]. Supporting this view, in vivo assays indicated that very low concentrations (0.01–2 μM) of As(III) and MMAs(III) inhibit the activity of PARP-1, which contains two C3H1-type ZnFs [111, 114,115,116]. Interestingly, it has been suggested that As(III) binding to the ZnFs of PARP-1 per se does not exert an inhibitory effect: instead, the As(III)-bound cysteines in PARP-1 are highly susceptible to oxidation by ROS, which ultimately leads to its inactivation [117, 118]. In addition, As(III)-induced ROS or RNS may also be responsible for oxidation/S-nitrosylation of PARP-1 resulting in Zn(II) loss and inhibition of its activity [119, 120]. PARP-1 acts as a sensor of SSBs and DSBs, which stimulate PARP-1-mediated synthesis of poly(ADP-ribose) (PAR) and the attachment of PAR chains onto itself and other proteins involved in the cellular response to DNA damage and DNA metabolism. The presence of PAR chains at the site of DNA damage facilitates repair processes by chromatin relaxation and by recruiting proteins involved in SSB repair, removal of bulky adducts by NER, removal of abortive TOP1cc, and DSB repair by HR and non-homologous end-joining (NHEJ) [121]. Inhibition of PARP-1 activity by 2 μM As(III) strongly perturbs the repair of H2O2- and ultraviolet radiation-induced DNA damage in human keratinocytes (HaCaT cells) [115, 116, 122]. Another function of PARP-1 is to downregulate repair of DSBs by HR and therefore, PARP-1 inhibition increases the level of sister chromatid exchanges [121], which is a hallmark of genomic instability frequently observed in human cells upon As(III) exposure [123]. Importantly, mouse embryonic fibroblasts lacking PARP-1 exposed to 11.5 μM As(III) exhibited elevated levels of DNA damage, micronuclei induction, telomere attrition and increased cell death [124]. This suggests that, in addition to inhibiting DNA repair, As(III) may itself induce DNA lesions that are repaired by PARP-1-dependent pathways.
The xeroderma pigmentosum complementation group A (XPA) protein, which contains the C4-type ZnF, is another As(III)-targeted DNA repair protein [111, 122]. XPA plays a central role in NER by coordinating the assembly of core NER factors at the site of DNA damage [125]. Initially, it was suggested that As(III) does not interfere with the binding of XPA to UV-irradiated oligonucleotides [126]. Another group reported that As(III) disrupts the binding of XPA to DNA oligomers crosslinked with mitomycin C only at very high concentrations (0.5–2 mM) [127]. Moreover, MMAs(III) was found to exhibit much higher affinity for the XPA ZnF compared to As(III) [128]. However, more recent reports demonstrated that As(III) at the concentrations of 0.5–2 μM was able to decrease XPA binding to chromatin in human keratinocytes (HEKn cells) [122] and XPA and PARP-1 displayed similar affinities for As(III) [129].
Finally, As(III) can bind to the RING finger domain of E3 ubiquitin ligases, such as RNF20–RNF40, FANCL, and RAD18, leading to inhibition of DSB repair, DNA interstrand crosslink repair (ICR), and replication bypass of UV-induced DNA lesions by translesion synthesis (TLS), respectively [130,131,132]. This suggests that As(III) may interfere with the function of RING-type E3 ubiquitin ligases in the cell, possibly disrupting not only DNA repair pathways but also ubiquitin-dependent protein degradation. As(III) binding to ZnF proteins may also stimulate their degradation, as shown for the histone acetyltransferase TIP60 in human HEK293T cells treated with 2–5 μM As(III) [133] and the oncogenic PML-RARα fusion protein in APL cells at the therapeutic concentration of 1 μM [37]. Notably, TIP60 activity is not only important for gene regulation but also for facilitating DSB repair by HR [134].
Less is known about the effect of Sb(III) on DNA repair. In A549 human lung adenocarcinoma cells, SbCl3 at subcytotoxic and cytotoxic concentrations (0.25–0.5 mM) was shown to specifically inhibit the repair of ultraviolet C (UVC) radiation-induced cyclobutane pyrimidine dimers by NER. This was associated with reduced expression of the NER protein XPE and the displacement of Zn(II) from XPA, which resulted in decreased binding of XPA to chromatin [135]. In vitro, Sb(III) binds to a CCCH-type peptide with high affinity, but also to CCHC-type ZnF peptides [136, 137]. Non-cytotoxic concentrations of Sb(III) (1–8 μM) have also been shown to trigger degradation of the RING finger PML-RARα oncoprotein in the APL cell line NB4 [138, 139]. Thus, similar to As(III), Sb(III) may have the potential to inhibit C3H1 and C4-type ZnF proteins involved in DNA repair. In line with this notion, an early report demonstrated that high concentrations of Sb(III) (0.2–0.4 mM) significantly inhibited DSB repair in Chinese hamster ovary cells (CHO-K1) [140].
More recently, it has been shown that non-cytotoxic concentration of Sb(III) (10 μM) also inhibited the repair of radiation-induced DSBs in human HeLa S3 cells in all phases of the cell cycle, suggesting that Sb(III) impairs DSB repair by both NHEJ and HR [91]. Importantly, Sb(III) did not interfere with the initial steps of DDR, but decreased the recruitment of the HR repair proteins BRCA1 and RAD51 to sites of DSBs [91]. BRCA1 is likely a direct target of Sb(III) due to the presence of a RING domain within BRCA1, and since As(III) has been reported to release Zn(II) from BRCA1 [132]. 0.2 mM Sb(III) also inhibited DSB repair in budding yeast by impairing NHEJ-dependent processes such as fusion of unprotected telomeres and re-joining of a linearized plasmid, and by negatively affecting the repair of phleomycin-induced DSBs [68].
The cytoskeleton as a target for arsenic and antimony cytotoxicity
It is well established that As(III) can cause aneuploidy, which is a driving force in many types of cancer [141, 142]. Early reports suggested that As(III) may induce chromosome missegregation by disrupting the mitotic spindle. 16 h exposure of mouse fibroblasts (Swiss 3T3 line) to high concentrations of As(III) (20 μM) resulted in morphological loss of microtubules [107], while low concentrations of As(III) (0.01–0.1 μM for 24 h) distorted the microtubules of the spindle apparatus and induced aneuploidy in human lymphocyte cultures [143,144,145]. As(III) also inhibited guanosine triphosphate (GTP)-induced polymerization of tubulin at 1 mM, while MMAs(III) and DMAs(III) caused inhibition at 10 μM, as demonstrated by the acellular tubulin assembly assay [146]. A more recent study reported that 20–40 μM As(III) inhibited the in vitro assembly of microtubules and a significant number of rat lung fibroblasts (RFL6 cell line) exposed to 10 μM As(III) for 24 h exhibited damage to spindle microtubules [147]. In mammalian cells, As(III) has been proposed to interfere with tubulin polymerization and microtubule formation by binding to two vicinal cysteine residues (Cys12 and Cys213) within the β-tubulin monomer, thereby preventing GTP binding, which is indispensable for the formation of tubulin polymers [145]. In contrast, G2 phase-enriched human fibroblasts exposed to 5 μM As(III) for 24 h exhibited derangement of the spindle apparatus and chromosome loss, but no inhibition of spindle formation was observed [148]. Similarly, another study found that disruption of mitosis by 5 μM As(III) in three human cell lines was not dependent on direct inhibition of tubulin polymerization but instead involved heat shock-like perturbation of centrosome function, resulting in centrosome fragmentation, and multi-polar spindle and mitotic arrest [149]. In budding yeast, subtoxic concentrations of As(III) [150, 151] and Sb(III) [68] also distort the morphology of the microtubule cytoskeleton, likely by inhibiting the chaperonin TRiC/CCT complex required for tubulin folding but not by inhibiting tubulin polymerization [151]. However, it is important to emphasize that yeast tubulin lacks the Cys12 residue, which was proposed to be targeted by As(III) in human tubulin. Perturbation of the spindle assembly checkpoint (SAC) resulting in metaphase arrest bypass is another possible mechanism of As(III)-induced aneuploidy [141]. For example, arsenic-induced squamous cell carcinomas are characterized by increased expression of microRNA-186, which negatively modulates expression levels of SAC regulators [152]. Moreover, human keratinocytes overexpressing microRNA-186 exhibited increased levels of aneuploidy during chronic exposure to low concentrations of As(III) (100 nM for 8 weeks) [152, 153].
Like tubulin, actin has been identified as an As(III)-binding protein in human cells [145, 154]. As(III)-induced disruption of the actin cytoskeleton was demonstrated at 2.5 μM in mouse fibroblasts (Swiss 3T3 line) [143,144,145], 5 μM in human acute leukemia cells (HL-60 line) [155] and 10 μM in mouse endothelial cells (SVEC4-10 line) [156]. In budding yeast, both As(III) [150, 151] and Sb(III) [68] at subtoxic concentrations also triggered reorganization of the actin cytoskeleton. Similar to tubulin, it has been suggested that As(III) interferes with actin folding by inhibiting the chaperonin TRiC/CCT complex [151]. Interestingly, other stress conditions, including oxidative stress, cause similar rearrangements of the actin cytoskeleton that may result from the formation of an intramolecular disulfide bond between conserved Cys374 and Cys285 residues or an intermolecular disulfide bond with Cys374 of another actin molecule [157, 158]. Importantly, the yeast act1-Cys285A,C374A mutant failed to reorganize the actin cytoskeleton in response to acute oxidative stress and exhibited increased sensitivity to oxidative stress [157]. This suggests that stress-induced rearrangement of the actin cytoskeleton may serve as an adaptive/protective stress response. Indeed, in yeast cells, As(III)-induced depolarization of the actin cytoskeleton was transient, followed by a recovery of the polar actin distribution accompanied by resumption of growth [150, 151]. The phenotype of yeast cells expressing the mutant actin lacking Cys285 and Cys374 in the presence of As(III) and Sb(III) has not been reported.
There is increasing evidence pointing to an important contribution of nuclear actin filaments in DSB repair by modulating chromosome movements and clustering of repair sites [159]. On the other hand, the interphase cytoplasmic microtubule network participates in trafficking of DNA repair factors to the nucleus [160] and facilitates DNA repair by promoting changes in nuclear morphology and chromatin organization as well as mobility of DNA lesions in mammalian cells [161]. Whether As(III) and Sb(III) exert their negative effects on DNA repair by disrupting nuclear actin filaments or interphase cytoplasmic microtubules, remains to be investigated both in yeast and human cells.
In conclusion, both As(III) and Sb(III) appear to exhibit similar complex and indirect genotoxicity mechanisms, including replication-associated DNA damage mediated by oxidative stress and DNA–protein crosslinks as well as telomere dysfunction, which are exacerbated by metalloid-induced inhibition of key DNA repair pathways. Moreover, recent data obtained in a yeast model suggest that As(III) and Sb(III) may also have the ability to damage DNA independently of oxidative stress, replication or DNA–protein adducts. Although genotoxicity of As(III) at cytotoxic concentrations is evident in in vitro studies and in exposed humans, less is certain with regards to how As(III) contributes to carcinogenesis during long-term exposure to environmental relevant concentrations. As(III)-induced aneuploidy, which is also observed at low concentrations of As(III), may be another driving force of As(III)-induced genomic instability and also proteome imbalance. Low doses of As(III) inhibit expression and activity of key proteins involved in genome maintenance and cell cycle regulation, such as PARP-1 [111, 114,115,116] and p53 [162, 163]. In addition, different cell types or ages may vary in their telomere length, as well as their antioxidant, DNA repair, arsenic methylation and detoxification capacities, which may greatly influence susceptibility to As(III)-induced malignant transformation. Given the strong co-carcinogenic properties of As(III) and environmental pollution, co-exposure to other toxic metalloids and heavy metals, such as antimony and cadmium, and other pollutants, is also expected to promote carcinogenesis. Finally, it has recently been revealed that As(III) exhibits immunotoxic and immunosuppressive effects [164]. Some of these mechanisms may also be relevant for Sb(III), but both the genotoxic and carcinogenic properties of this metalloid require further extensive research.
How arsenic causes proteotoxicity
Proteotoxic stress has emerged as an important contributor to the toxicity of arsenic. Traditionally, As(III) toxicity has been attributed to its interactions with sulfhydryl groups in folded native proteins, altering their function, regulation, and interactions [33, 36]. Recent studies revealed an additional mode of toxic action in which As(III) targets non-native proteins, thereby impairing their proper folding. Specifically, As(III) was shown to interfere with the refolding of chemically denatured proteins in vitro, with protein folding in vivo, and to cause misfolding and aggregation of nascent proteins in living cells.
Protein folding and quality control is crucial for cell physiology and survival. To perform their biological function, most proteins must first adopt their native conformation, i.e., their folded three-dimensional structure. Protein folding takes place on the ribosome during translation, in the cytoplasm after ribosomal release, or in organelles such as the endoplasmic reticulum and mitochondria. If the correct native fold is not attained or lost, proteins can misfold and aggregate. Protein misfolding can occur due to mutations, external stress conditions, errors during transcription and translation, and during cellular aging [165, 166]. Protein misfolding and aggregation is detrimental for cells and organisms and is a feature of many human diseases including metabolic, oncological, and neurodegenerative disorders [167]. To ensure a functional proteome (protein homeostasis or proteostasis), cells use protein quality-control (PQC) systems (Fig. 2) that encompass (a) molecular chaperones that assist in the folding of proteins into their native conformation and in the disaggregation, refolding, sequestration, and degradation of non-native proteins; and (b) protein degradation systems such as the ubiquitin–proteasome system (UPS) and autophagy-lysosome pathway that eliminate misfolded and aggregated proteins [166, 168,169,170].
How arsenite causes proteotoxicity. A As(III) may bind to free thiols or other functional groups in nascent and non-native proteins, thereby preventing their folding into the native conformation, promoting protein misfolding and aggregation. As(III) may also impair chaperone-mediated folding and disaggregation by binding to the substrate protein, to molecular chaperones, and by modifying the structure of the aggregate. B The protein aggregates formed during As(III) exposure may contribute to toxicity by provoking aberrant protein–protein interactions, by sequestering chaperones, and by increasing the misfolding of other proteins that have not encountered the metalloid. Additionally, As(III) can affect aggregate structure such that processing by chaperones and possibly other PQC factors is impaired. The figure was created with BioRender.com
This section will highlight how protein aggregates are formed during As(III) stress, how these aggregates cause toxicity, and how cells maintain a healthy proteome during As(III) exposure. Most of the studies on metalloid-induced proteotoxicity described below have been performed in vitro and in model systems, primarily budding yeast and various human cell lines, whilst mechanistic studies in native cells and tissues are scarce.
Misfolding and aggregation of nascent and non-native proteins is the prime mechanism of arsenite-induced proteotoxicity
A number of in vitro studies demonstrated that As(III) disrupts protein folding at µM concentrations. Initial studies indicated that the binding of As(III) to cysteine-containing model peptides perturbed their structures [171, 172]. Follow-up studies showed that As(III) inhibits oxidative refolding of certain disulfide-containing proteins (lysozyme, ribonuclease, riboflavin-binding protein) by binding to cysteine residues within the reduced and unfolded proteins [173, 174]. As(III) also interfered with spontaneous refolding of chemically denatured luciferase that possesses four cysteine residues but no disulfide bridge. Chaperone-mediated refolding of luciferase was also inhibited by As(III), but its impact was greater on the substrate than on the chaperone system [175]. Importantly, the defect in chaperone-mediated refolding of green fluorescent protein was abrogated in a cysteine-free version of the substrate protein [176], providing strong evidence that As(III) directly modifies cysteine residues in non-native target proteins. Together, these in vitro studies established that As(III) can inhibit the formation of the native protein fold by forming complexes with cysteine side chains (Fig. 2A).
As(III) has a strong impact on protein folding also in vivo. Studies in S. cerevisiae demonstrated that As(III) at concentrations ranging from 100 µM (subcytotoxic) to 1.5 mM (cytotoxic) provokes misfolding and aggregation of hundreds of cytosolic proteins and that aggregation is concentration-dependent [175,176,177,178]. Intracellular As(III) appears to be the culprit since overexpression of an As(III) export protein not only decreased intracellular arsenic concentrations but also protein aggregation levels [175]. Similarly, loss of yeast proteins that either restrict or enhance cytosolic arsenic concentrations is accompanied by reduced or increased protein aggregation levels, respectively, during As(III) stress [178].
What proteins are targeted by As(III) for misfolding and aggregation? Several observations indicate that As(III) primarily targets nascent or non-native proteins for misfolding and aggregation (Fig. 2A). In vitro experiments showed that As(III) has a modest impact on the activity of native luciferase whereas spontaneous and chaperone-mediated luciferase refolding were strongly inhibited [175]. In vivo experiments in yeast cells revealed that newly synthesized proteins aggregated during As(III) stress, and that As(III)-induced protein aggregation at concentrations of 100–500 µM was supressed when protein biosynthesis was inhibited, either chemically using a translation inhibitor [175] or genetically by deleting genes encoding proteins with functions in cytosolic translation [178, 179]. Proteomic studies further indicated that yeast proteins that are susceptible for aggregation during exposure to 1.5 mM As(III) have high translation rates and are substrates of ribosome-associated Hsp70 chaperones [175, 177, 180] while they appear relatively stable in their native (folded) state [177]. Collectively, these findings indicate that As(III) primarily impairs folding of nascent proteins rather than inducing large-scale unfolding of native proteins.
In addition to acting on non-native proteins, As(III) may also impair chaperone-assisted protein folding and disaggregation (Fig. 2A). In vitro experiments with the Hsp70 chaperone system of Escherichia coli (DnaK, DnaJ, GrpE) showed that As(III) interferes with chaperone-assisted refolding of denatured and heat-aggregated luciferase, even though the main impact of As(III) was on the substrate rather than on the chaperone system [175]. As(III) also inhibits substrate folding by the bovine and archaeal chaperonin TRiC/CCT in vitro [151]. As(III) was shown to bind to individual members of the human TRiC/CCT complex using a human proteome microarray [181] as well as to actin and tubulin in human lymphoblastoid cells and human breast cancer cells [145, 154] that are major TRiC/CCT substrates. Thus, it is not clear whether As(III) primarily affects TRiC/CCT activity or the folding substrate. Interestingly, the purified yeast Hsp70 and bichaperone Hsp70–Hsp104 systems were inhibited by As(III) to similar degrees as the bacterial Hsp70 system [175, 176]. However, instead of inhibiting the yeast chaperones, As(III) impaired protein disaggregation by modifying the structure of the aggregates, thereby preventing efficient binding of the chaperones to the aggregate [176]. One way As(III) could affect aggregate structure is by acting as an intermolecular crosslinker between two or three polypeptide chains leading to the formation of heterodimers and trimers [32, 36, 176]. Possibly, the observed changes of aggregate properties in the presence of As(III) [176] may not only reduce chaperone binding but might also affect aggregate processing by other PQC systems.
Experimental data support the notion that folding inhibition is an important contributor to the toxicity of As(III) that acts in parallel with other toxicity mechanisms. A genome-wide screen found a strong correlation between yeast mutants with enhanced protein aggregation levels and As(III) sensitivity, and between mutants with reduced aggregation levels and As(III) resistance [178]. Likewise, several studies in different model systems, including yeast (500 µM–1.5 mM), worm (Caenorhabditis elegans) (1.5–4.5 mM) and various mammalian cell lines (10–100 µM), indicated that maintaining proteostasis is crucial for cell survival and proliferation during As(III) exposure [175, 176, 178, 179, 182,183,184,185,186,187]. How aggregates formed during As(III) stress affect cell viability is not well understood. Protein aggregates can cause toxicity through multiple mechanisms. For example, aggregation could result in loss-of-function of individual proteins with protective or detoxification functions. Aggregation could also create a toxic gain-of-function where aggregated proteins damage membrane lipids, interact inappropriately with proteins and RNA, and sequester molecular chaperones and PQC factors [166]. Most likely, several mechanisms contribute to the toxicity caused by the aggregates formed during As(III) stress (Fig. 2B): (a) proteins that aggregate during As(III) stress are enriched for proteins with many interaction partners, suggesting that misfolded forms of these proteins could be involved in aberrant interactions [177]; (b) multiple chaperones associate with the aggregated proteins [175, 177], suggesting that chaperones may be sequestered away from the active pool of cytosolic PQC factors during As(III) stress; (c) in vitro experiments suggest that As(III)-aggregated protein seeds can drive misfolding and aggregation of other proteins that have not encountered the metalloid [175]; (d) aggregate size and structure may influence their seeding capacity and toxicity [166], and As(III) has been shown to modify the structure of aggregated model proteins [176, 188]. It will be important to elucidate the structure and properties of aggregates formed in the presence of As(III) to better understand their in vivo toxicity.
How does As(III) inhibit the folding of non-native proteins in vivo? On the molecular level, As(III) probably acts on not-yet folded segments of the polypeptide chain emerging from the ribosome during protein synthesis, preventing the formation of the native protein conformation. As(III) can bind to one, two or three thiol groups of cysteine residues with unidentate binding being weak whereas tridentate binding is highly stable [36]. Formation of stable pluridentate complexes between As(III) and exposed side chains is more likely to occur during the folding process when the protein backbone is flexible, and their formation prevents the protein from adopting its native conformation, promoting its misfolding and aggregation. In contrast, side chains are more likely to be fixed or buried inside the folded native protein, implying that formation of pluridentate complexes would require partial protein unfolding. We anticipate that As(III) affects folding of most nascent and non-native cellular proteins, and its impact on folding is likely influenced by the concentrations of both As(III) and the target proteins, the amino acid sequence of the target protein, the content and arrangement of thiol groups in side chains, the folding pathway, the rate of synthesis and folding, the structure of folding intermediates, and the efficacy of As(III) detoxification and PQC systems. While As(III) primarily targets cysteine-containing non-native proteins for aggregation, aggregation might also involve intermolecular hydrophobic interactions between As(III)-misfolded proteins and proteins that have not encountered the metalloid [32]. The observations that As(III)-aggregated proteins are enriched in multiple protein–protein interactions [177] and have seeding capacity [175], support this notion.
Additional ways arsenic could affect proteostasis and induce proteotoxicity include depletion of cellular ATP. In yeast, this mechanism appears unlikely for As(III), since As(III) concentrations that results in widespread protein aggregation in vivo (500 µM) had a negligible effect on intracellular ATP concentrations [176]. However, since ATP depletion can affect proteostasis [189], it is possible that As(V) could induce protein aggregation through disruption of ATP generation, although this remains to be demonstrated. Errors during transcription and translation may also lead to protein misfolding and aggregation [190, 191]. However, in budding yeast, 500 µM As(III) did neither induce errors during transcription [178], nor did it enhance mRNA mistranslation [175]. Finally, As(III) could affect proteostasis not only by stimulating the formation of protein aggregates, but also by interfering with their clearance through degradation. Studies in various mammalian cell lines showed that low As(III) concentrations (0.25–2 µM) can inhibit autophagic flux by blocking the autophagosome-lysosome fusion step through inhibition of SNARE complex formation [192, 193]. Whether this inhibition leads to the build-up of aggregates remains to be demonstrated. As(III) can bind to E3 ubiquitin ligases with RING finger domains [33, 194] and some studies using NIH3T3 and HEK293 cells suggested that arsenic (50–250 µM As(V), 1–10 µM As(III)) may inhibit the UPS [195, 196]. However, direct measurements of proteasomal activity revealed that, in fact, it increases in As(III)-exposed yeast (500 µM) and HeLa (25 µM) cells [175, 185, 197]. Nevertheless, it cannot be excluded that changes of aggregate structure by As(III) [176, 188] could affect aggregate processing and degradation.
Taken together, the impact of As(III) on non-native proteins, on chaperone machineries, and on aggregate structure and processing is expected to result in substantial protein misfolding and aggregation in cells, placing a heavy burden on the PQC systems.
Do other metals and metalloids including antimonite disrupt protein folding in cells?
We predict that many more metals and metalloids have the potential to disrupt protein folding processes in living organisms [32]. Indeed, like As(III), cadmium [Cd(II)] induces misfolding and aggregation of nascent and non-native proteins in vivo. Interestingly, Zn(II) mitigated Cd(II)-induced protein aggregation in yeast cells [198], suggesting that Zn(II) may be substituted by Cd(II) while the nascent polypeptide chain undergoes the folding process [198, 199]. In contrast, Zn(II) did not alleviate As(III)-induced protein aggregation in yeast cells [198] even though As(III) can bind to ZnF motifs [37, 111]. Copper [Cu(I)] was shown to cause widespread protein aggregation in E. coli, probably by targeting cysteine- and histidine-containing proteins [200]. Whether Cu(I) primarily targets native or non-native proteins, remains to be determined. In case of chromium [Cr(VI)], protein aggregation in budding yeast is caused by enhanced mRNA mistranslation [201], but the underlying mechanism remains unknown. In vitro studies showed that mercury [Hg(II)] and lead [Pb(II)] can interfere with the refolding of chemically denatured proteins [202], but their effects on protein folding in vivo is not known. As outlined above, Sb(III) has high affinity for thiols and its toxicity may, in part, be attributed to its capacity to bind to cysteine residues in proteins [15, 42, 43, 136, 137]. Thus, it is conceivable that Sb(III) could disrupt protein folding in cells. A few studies suggest that Sb(III) might induce proteotoxic stress in cells: the molecular chaperones HSP70 and HSC70 were shown to confer Sb(III) tolerance to the protozoan parasite Leishmania [203] and work in yeast implicated the UPS in the protection from Sb(III) toxicity [182]. An in vitro study showed that Sb(III) can interact with and induce conformational changes in bovine serum albumin and to promote its partial aggregation [204]. Finally, antimony trioxide at high concentrations (400 µM–2.8 mM) induced transcription of genes encoding molecular chaperones as well as UPS- and autophagy-related genes in Daphnia magna [205]. Nevertheless, direct evidence that Sb(III) affects protein folding is currently lacking, which calls for more research in this area.
Arsenite induces the formation of biomolecular condensates
As described above, As(III) can induce the formation of protein aggregates that are largely insoluble assemblies of misfolded proteins with aberrant and non-native conformations. Additionally, As(III) has been implicated in the formation of certain biomolecular condensates that are membraneless assemblies of proteins and nucleic acids. In contrast to aggregates, biomolecular condensates are characterized by weak, dynamic and multivalent interactions, are often formed via liquid–liquid phase separation, and carry out specialized functions in eukaryotic cells during physiological and stress conditions [206, 207]. Examples of condensates and their functions include stress granules (mRNA storage transcriptional regulation), processing bodies (mRNA decay and silencing), promyelocytic leukemia nuclear bodies (apoptotic signaling, anti-viral defence, DNA repair and transcription regulation), actin patches (endocytosis), heterochromatin (gene regulation), and nucleolus (ribosomal synthesis). Protein condensation and aggregation appear to be interlinked: (a) some condensates can transition from liquid-like states into more solid-like states when misfolded proteins associate with the condensates, (b) the cellular PQC machinery is implicated in the regulation of condensate formation, dissolution and dynamics, (c) a decline in PQC may contribute to the formation of aberrant, disease-causing condensates, and (d) there is evidence that certain condensates can serve as PQC compartments themselves [206, 208].
High concentrations of As(III) stimulates the formation of stress granules (SGs) and processing bodies (PBs) [209, 210] that are dynamic and reversible biomolecular condensates involved in mRNA storage and processing, and in transcriptional regulation [211]. Formation of SGs correlate with As(III)-induced translation repression through phosphorylation of the key translation initiation factor eIF2α [209, 212] and SG formation is important for stress adaptation [206, 211]. In contrast, PB formation does not involve eIF2α phosphorylation [209]. Work in S. cerevisiae suggests that the protein aggregates formed during As(III) stress are largely distinct from PBs and SGs as their localization and protein composition appear to differ to a large extent [175, 178]. Nevertheless, the PQC machinery is not only implicated in the processing of misfolded proteins but also in the regulation of SG and PB formation and dissolution, indicating cross-talk between these structures [206, 213].
Promyelocytic leukemia nuclear bodies (PML-NBs) are membraneless structures inside the nucleus involved in multiple genome maintenance pathways including the DNA damage response, DNA repair, telomere homeostasis, and p53-associated apoptosis [214]. The PML protein is the key driver of the formation of PML-NBs. The PML protein is also involved in the pathogenesis of APL and, as outlined above, arsenic trioxide can cure APL by binding to cysteine residues in the PML-RARα fusion protein inducing its oligomerization and subsequent degradation [37, 38]. Interestingly, As(III) stimulates PML-NB formation followed by PML degradation upon extended exposure in non-APL (HeLa) cells [215]. Additionally, PML-NBs have been proposed to have a function in nuclear protein homeostasis: upon proteotoxic stress, PML-NBs compartmentalize defective proteins and recruit chaperones and proteasomes to promote efficient degradation [216]. It is tempting to speculate that PML-NBs constitute a nuclear PQC compartment also during As(III) stress. In such a scenario, PML might act as an As(III) sensor that stimulates NB formation followed by recruitment of misfolded proteins and PQC components to restore proteostasis.
Other examples of condensates where misfolded proteins are stored, refolded or degraded during cellular stress conditions include the insoluble protein deposit (IPOD) and the juxtanuclear protein quality control (JUNQ) compartment [169, 206]. If and how As(III)-misfolded are partitioned into specific subcellular sites remains to be explored.
Recent studies indicate that DSB repair involves condensate formation [217]. Induction of DSBs triggers a local accumulation of DNA damage signaling and DNA repair proteins, called DDR foci, at sites of DNA breaks. Proper assembly and disassembly of DDR foci is crucial for rapid and efficient DNA repair coordinated with cell cycle arrest followed by recovery. Several DDR proteins have been shown to exhibit features of phase separation and to form condensates, such as the HR factor Rad52 in S. cerevisiae [218, 219] and the DNA damage signaling protein and DNA repair pathway choice regulator 53BP1 [220, 221] in human cells. It is believed that condensate formation may facilitate tethering of DNA molecules and control the recruitment and accumulation of DNA repair proteins at sites of DSBs or exclusion of other proteins [222]. For example, Rad52 condensates mediate nucleation of DNA damage-inducible intranuclear microtubule filaments, which facilitate Rad52 condensate fusion resulting in clustering of DSBs at the nuclear periphery for repair [218]. Interestingly, proteins condensates have also been implicated in polymerization, organization and dynamics of both tubulin and actin cytoskeleton [223,224,225]. 53BP1 condensates act as a scaffold for the tumor suppressor protein p53, promoting its activation, thereby inducing changes in transcription of p53-targeted genes and checkpoint activation in response to DNA damage [220]. On the other hand, 53BP1 is excluded from the PAR-seeded condensates of the low complexity domain-containing FET proteins (FUS, EWS, TAF15) [226]. FET condensates are an early but transient event at DNA damage sites and can therefore control the spatiotemporal assembly and disassembly of DDR proteins. A recent in vitro study demonstrated that telomeres also undergo a phase separation-driven compartmentalization that regulates the access of telomere-associated and DNA repair proteins to chromosome ends [227]. Considering the observed negative effects of As(III) and Sb(III) on DSB repair, PARP-1-mediated PAR synthesis, cytoskeleton morphology and telomere stability, it is tempting to speculate that the mechanisms of metalloid toxicity may also involve disruption of biomolecular condensate-driven cellular processes by interfering with post-translation modifications and/or folding of proteins involved in condensate formation.
Regulation of eukaryotic gene expression has been shown to involve condensate formation of gene-specific transcription factors with coactivator and RNA polymerase II complexes [228,229,230]. Whether metalloids promote condensate formation of specific transcription factors involved in detoxification and defence remains unknown.
To conclude, recent studies established that the prime mechanism of As(III)-induced proteotoxicity is governed by interactions between As(III) and exposed cysteine residues in nascent or non-native proteins. This interaction obstructs the formation of the native protein conformation and promotes protein misfolding and aggregation. Additionally, As(III) may potentially regulate or disrupt formation of biomolecular condensates as a toxicity or defence mechanism. Due to the importance of biomolecular condensates in health and disease, future research efforts should be directed toward the elucidation of how arsenic, antimony and other metals affect condensate biogenesis.
Mechanisms that protect cells from metalloid-induced protein misfolding and aggregation
Cells rely on two main strategies to maintain a functional proteome during As(III) exposure. The first strategy relies on damage prevention. Cells can protect nascent proteins from harmful arsenic interactions by limiting intracellular arsenic concentrations. Yeast cells respond to As(III) by regulating influx, efflux, and sequestration systems [46, 51, 231,232,233,234], and this response is important to safeguard proteostasis: yeast cells capable of restricting cytosolic arsenic concentrations show reduced protein aggregation levels whilst a failure in limiting cytosolic arsenic is accompanied by enhanced protein aggregation [178]. Regulation of certain arsenic influx and efflux systems involves the transcription factor Yap8 and the MAP kinase Hog1 that act as direct sensors of intracellular As(III) [39, 41, 51]. Thus, these proteins couple arsenic-binding to improved proteostasis and cell survival. Intracellular As(III) chelation by GSH also protects cells from extensive protein aggregation, probably by lowering the cytosolic concentration of ‘free’ As(III) that can interfere with protein folding processes [233].
Another way of preventing or reducing aggregate formation is to repress translation. A number of studies in yeast (500 µM–1.5 mM) and human cell lines (10–150 µM) demonstrated that As(III)-exposed cells repress global protein biosynthesis, probably as a result of increased phosphorylation of the translation initiation factor eIF2α and subsequent inhibition of translation initiation [178, 179, 183, 187, 235, 236]. Additionally, studies in yeast cells showed that expression of genes encoding aggregation-prone proteins [177] and of protein biosynthesis–related genes [183, 234, 237] were repressed in response to As(III) exposure. Translation repression is crucial for maintaining proteostasis during As(III) stress, since yeast mutants with low global translation activity as well as mutants with improved translation repression efficiency are less susceptible for As(III)-induced protein aggregation and toxicity [178, 179]. Conversely, yeast mutants that are defective in translation repression show enhanced protein aggregation levels and As(III) sensitivity [178]. The importance of translation control during As(III) stress is further emphasized by the observation that deletion of yeast genes encoding functions in protein biosynthesis leads to reduced protein aggregation and As(III) resistance [151, 178, 179, 183]. Together, these studies establish that an accurate control of protein synthesis is of central importance to ensure proteostasis and cell viability during As(III) stress, probably by lowering the influx of misfolded proteins into the proteome. Little is known about the sensing and signaling mechanisms that regulate translation during As(III) stress, although the heme-regulated inhibitor kinase (HRI) in mouse cells [187] and the TORC1 (TOR complex 1) protein kinase in yeast [238] have been implicated.
The second strategy relies on damage elimination. Studies in various cell types and model organisms have shown that expression of genes encoding functions in protein folding and degradation is induced in response to As(III) (concentrations ranging from 0.5 to 50 µM for mammalian cells; 0.5–1.5 mM in yeast) [46, 175, 185, 196, 234, 237, 239,240,241,242]. Expression of genes related to ATP production is also induced by As(III) [234, 237], and yeast cells ensure that sufficient ATP is available for their protein folding and degradation needs [176]. Protein aggregates formed during As(III) stress are preferentially degraded through the UPS (Fig. 2A). Studies in budding yeast showed that the aggregates are tagged with K48-linked ubiquitin chains [176, 179] and that cells increase the amount of proteasomal components as well as proteasomal activity during As(III) stress [175, 183, 234, 237]. Inhibition of proteasomal activity by genetic or chemical means impairs the clearance of aggregates [175, 176, 178] and sensitizes yeast cells to As(III) [175, 176, 182, 183]. Similarly, mouse fibroblasts and HeLa cells exposed to 25 µM As(III) accumulate K48-linked ubiquitin conjugates and increase proteasomal activity [185, 197]. Interestingly, inhibition of protein ubiquitination abrogates As(III)- and heat-induced stimulation of proteasomal activity in HeLa cells, suggesting that proteasomal activation may be signaled by the build-up of ubiquitinated proteins in the cells [197]. Studies in mouse cells and C. elegans indicated that proteasomes may adapt to As(III) stress via the arsenite-inducible RNA-associated protein (AIRAP), that is induced by As(III) and associates with proteasomes to regulate their degradative capacity [185]. Yeast cells also possess As(III)-inducible components of the proteasome including Cuz1 (ZFAND1 in mammalian cells) and Tmc1 (an AIRAP orthologue). Yeast Cuz1 and mammalian ZFAND1 interact with the proteasome and with Cdc48 (p97 in mammalian cells), an ATPase that delivers proteins to the proteasome for degradation, while Tmc1 and AIRAP associate with the proteasome’s 19S cap [182, 243, 244]. Loss of either Cuz1 or Tmc1 sensitizes yeast cells to As(III) and Sb(III) [182]. Similarly, loss of AIRAP sensitizes worms to As(III) [184]. Whether cells lacking these regulators accumulate protein aggregates during As(III) stress remains to be determined. Moreover, it is currently not known how these proteins regulate proteasomal activity during As(III) stress or if and why they are specifically required during proteotoxic stress caused by As(III). One possibility is that changes in aggregate structure inflicted by As(III) [176, 188] necessitates an adaptation of the proteasome for efficient substrate degradation. In addition to K48-linked chains, the aggregates formed during As(III) stress are also tagged with K63-linked chains [179]. K63-linked chains have primarily been associated with proteasome-independent processes such as DNA repair, endocytosis and selective autophagy [245, 246] but have recently been shown to play a role in proteasome-dependent substrate degradation when forming branched K48/K63 chains [247, 248]. Since branched ubiquitin chains are stronger degradation signals than unbranched chains [247, 248], the presence of both K48- and K63-linked chains on As(III) aggregated proteins raises the possibility that these aggregates may be difficult substrates for the proteasomes. Nevertheless, whether the K48 and K63 chains assembled on As(III)-aggregated proteins are homotypic or heterotypic branched K48/K63 chains remains to be demonstrated. Similarly, the ubiquitin ligases that mark the proteins that misfold during As(III) stress with K48 and K63-linked chains, remain to identified.
In addition to the UPS, the autophagy–lysosome pathway also contributes to aggregate clearance and As(III) resistance (Fig. 2A). Autophagy is activated in yeast (0.5–1.5 mM) and human bronchial epithelial cells (0.25 µM) exposed to As(III) [183, 249] and several yeast mutants lacking autophagy–lysosome pathway components have increased levels of protein aggregates during As(III) stress [176, 178]. While these findings implicate autophagy in the clearance of As(III)-induced aggregates, autophagy appears less prominent than the UPS [176, 183]. The weaker contribution of autophagy to aggregate clearance might be due that fact that As(III) can inhibit autophagic flux [192, 193] or that the two pathways recognize distinct subsets of misfolded and aggregated substrates.
The cellular concentration of molecular chaperones increases in response to As(III). Molecular chaperones are implicated in the folding, sorting, disaggregation, and degradation of proteins [166, 168, 169] implying that their upregulation is important to meet an increased protein folding, sorting, and/or degradation demand. Several chaperones co-sediment with aggregated proteins during As(III) stress in vivo [175], suggesting that they are engaged in the disaggregation, refolding or degradation of substrate proteins. Alternatively, these chaperone interactions might be important to prevent As(III)-misfolded proteins from erroneous interactions with other proteins. Chaperone-mediated disaggregation has been implicated in aggregate clearance: deletion or chemical inhibition of the yeast disaggregase Hsp104, its cytosolic co-chaperones Ydj1 (Hsp40) or Ssa1 and Ssa2 (Hsp70) delayed the clearance of protein aggregates formed during As(III) stress [176]. The fate of the proteins recovered from the aggregates is, however, not entirely clear. Heat-aggregated proteins are preferentially refolded rather than degraded in yeast [250] and thermotolerance is strongly dependent on Hsp104 [251]. In contrast, yeast cells lacking Hsp104 are not sensitive to As(III) at concentrations that provoke protein misfolding and aggregation [176, 252]. The observation that aggregates formed in the presence of As(III) are poor substrates for chaperone binding [176] might explain why Hsp104 is largely dispensable for growth and survival during As(III) stress. Moreover, overexpression of Hsp104 sensitizes yeast cells to As(III) [176], possibly due to high unfolding activity [253] and subsequent build-up of misfolded proteins [176]. These observations suggest that the prime fate of As(III)-misfolded proteins might be their degradation rather than refolding. Indeed, yeast Ydj1 and Ssa1/Ssa2 are not only involved in protein folding but also in protein turnover, keeping misfolded substrates soluble until they are ubiquitinated by ubiquitin ligases for subsequent degradation [254,255,256,257]. The GimC/prefoldin chaperone complex is also implicated in aggregate clearance during As(III) stress in yeast [179]. Similar to Hsp40 and Hsp70, GimC/prefoldin is primarily involved in nascent protein folding [258] but has also been implicated in protein turnover, facilitating degradation by maintaining substrate solubility [259]. Thus, molecular chaperones may be important to keep As(III)-misfolded proteins soluble, promoting their ubiquitination and degradation. Molecular chaperones may also direct misfolded proteins to specific subcellular sites, such as the IPOD and JUNQ, as a means to protect the intracellular environment [166, 169, 260,261,262,263]. Whether the aggregates formed during As(III) stress are sequestered to specific sites within the cell remains unknown.
Metalloid-induced genotoxicity and proteotoxicity: a double-edged sword targeting neurodegeneration
A common denominator of many age-related and neurodegenerative diseases is the dysfunction of proteostasis and the pathological accumulation of protein aggregates. Proteins that adopt non-native conformations in neurodegenerative diseases include amyloid β (Aβ) and tau in Alzheimer’s disease (AD), α-synuclein (αSyn) in Parkinson’s disease (PD), TAR DNA binding protein 43 (TDP-43) and RNA/DNA-binding protein fused in sarcoma (FUS) in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), and huntingtin (HTT) in Huntington disease (HD) [167, 264, 265]. Epidemiological studies have indicated an association between arsenic exposure and the prevalence of several neurodegenerative disorders, including AD and PD [13, 266,267,268,269,270]. However, the mechanisms by which arsenic contribute to these proteinopathies are poorly understood. Arsenic is a well-established neurotoxin that impairs cognitive functions and memory, especially in children, deteriorates mental health, and causes peripheral neuropathy. Several of these effects have been replicated in rodent studies [271, 272]. At the molecular level, it is believed that the neurotoxic properties of As(III) mainly stem from mitochondrial dysfunction and oxidative stress-derived cellular damage, disruption of proteostasis, neuroinflammation and neuronal apoptosis [13, 268]. Arsenic (and perhaps also antimony) can induce proteotoxicity and several yeast proteins that aggregate in As(III)-exposed cells have human or mouse orthologues that are implicated in proteinopathies and/or co-aggregate with disease-associated proteins in AD, familial ALS or PD [177]. Thus, metalloid-induced and disease-associated protein aggregation may have some shared features.
PD is characterized by the accumulation of αSyn aggregates in the neurons of the substantia nigra pars compacta, leading to neuronal cell death and, consequently, neurodegeneration [273]. Studies in neuroblastoma cell lines, rats, and mice revealed a time-dependent accumulation and oligomerization/aggregation of αSyn during exposure to low As(III) concentrations (0.03–0.3 µM) [274, 275]. Chronic exposure of mice to environmental relevant (0.65–6.5 μM) and high (65 μM) concentrations of As(III) in drinking water caused a dose-dependent increase in the phosphorylation of leucine-rich repeat kinase 2 (LRRK2) and αSyn in different brain regions [276]. Both LRRK2 and αSyn phosphorylation are implicated in PD, and phosphorylation is known to modulate αSyn oligomerization and fibrilization, as well as the formation of Lewy bodies and neurotoxicity [277, 278]. Several metals can affect αSyn conformation upon binding [279,280,281,282] and in vitro data indicate that As(III) may be incorporated into and alter the structure of amyloid fibres formed by acetylated human αSyn [188]. The presence of As(III) also affects the intracellular distribution and clearance of αSyn aggregates and aggravates αSyn toxicity in yeast cells [188]. Together, these observations suggest that A(III) may influence αSyn conformation, phosphorylation, regulation, and toxicity.
AD is characterized by insoluble deposits of various Aβ peptides, generated by cleavage of the β-amyloid precursor protein (APP), and of phosphorylated tau protein [283, 284]. Exposure to 5 μM As(III) and 10 μM DMAs(III) results in elevated APP levels in cholinergic cells. While DMAs(III) also increased Aβ formation, Aβ levels were unchanged or even reduced during As(III) stress [285]. However, when combined with Cd(II) and Pd(II), 5–50 μM As(III) greatly increased Aβ formation in rats, which was accompanied by cognitive impairments [286]. Additionally, formation of Aβ aggregates was stimulated by chronic exposure to As(III) (40 μM) in a mouse model of AD [287]. Together, these studies indicate that As(III) has the capacity to induce Aβ aggregation. As(III) has been shown to activate the tau kinases, such as GSK3, ERK1/2, JNK, and CDK5 in human neuroblastoma SH-SY5Y cells (5–10 μM) [287, 288] and in the rat brain (intraperitoneal injections of 2.5 mg/kg body weight for 28 days) [287,288,289,290] and consequently leading to hyperphosphorylation of several tau residues, which are also hyperphosphorylated under pathological conditions. As(III) (1–20 µM) also increased the formation of tau aggregates in human neuroblastoma SH-SY5Y cells at concentrations of 5–10 μM [288], consistent with the view that tau hyperphosphorylation leads to the formation of oligomers and neurofibrils [291, 292]. Thus, As(III) may induce tau aggregation by affecting its phosphorylation state.
Not much is known about the neurotoxic potential of antimony. Recent studies in mice revealed that Sb(III) exposure (10–40 mg/kg body weight) resulted in impaired learning and spatial memory [293] and neurotoxicity via ROS-mediated autophagic death of neurons [294]. Interestingly, long-term exposure of mice to Sb(III) (10–20 mg/kg body weight) triggered tau hyperphosphorylation and accumulation of tau and Aβ deposits [295]. Hence, Sb(III) exposure might be a risk factor for AD and other neurodegenerative diseases.
The above studies suggest that metalloids like As(III) and Sb(III) may contribute to proteinopathies by inducing misfolding and aggregation of specific disease-associated proteins such as αSyn, Aβ, and tau, by interacting with the protein, by inducing post-translational modifications such as phosphorylation, and/or via other mechanisms such as induction of oxidative stress (Fig. 3). Moreover, the strong impact of As(III), and perhaps also Sb(III), on global protein misfolding that propels extensive protein aggregation in cells, may also play a part in the initiation or progression of degenerative diseases. These effects are likely more severe with increasing age when the efficacy of the PQC systems decline.
How metalloid-induced genotoxicity and proteotoxicity may be interrelated. Arsenic and antimony may induce genotoxicity through oxidative stress, inhibition of DNA repair, perturbation of telomere maintenance, cytoskeletal abnormalities, and epigenetic dysregulation. Arsenite (and perhaps also antimonite) may cause proteotoxicity through oxidative stress, protein misfolding and aggregation and defective aggregate processing. Metalloid-induced genetic alterations may generate proteotoxic stress whereas protein aggregation may lead to enhanced oxidative stress and sequestration of genome maintenance and protein quality-control (PQC) components. Thus, metalloid-induced genotoxicity and proteotoxicity may be interrelated, where loss of genome integrity amplifies the impact of metalloids on proteome integrity and vice versa. In this way, genome instability and proteome instability are interrelated and jointly contribute to neurodegeneration and carcinogenesis. The figure was created with BioRender.com
It is important to note that beside pathological protein aggregation and aberrant proteostasis, neurodegenerative disease can arise due to several other mechanisms including DNA and RNA defects, neuronal cell death, synaptic and neuronal network defects, cytoskeletal abnormalities, and altered energy homeostasis [265]. In recent years, it has become increasingly clear that genome instability is another important contributor to neurodegenerative disorders [296] as well as cancer [297] and aging [298]. For example, AD, PD, ALS, HD, and FTLD are not only characterized by the accumulation of pathological protein aggregates, but also by increased levels of DNA damage [299,300,301,302]. A growing body of evidence suggests that age-related mitochondrial dysfunction and increased oxidative stress, accompanied by compromised DNA repair, result in genome instability that may precede and/or exacerbate proteome instability in neurons [303,304,305,306,307]. Several genetic disorders caused by mutations in genes encoding DNA damage sensors and DNA repair proteins also lead to neurodegenerative pathologies [308, 309]. Furthermore, loss or inhibition of human DNA damage sensor proteins, such as ATM, ATR, and MRE11, as well as induction of genotoxic stress in form of topoisomerase-DNA adducts, result in widespread protein aggregation [310,311,312]. Similarly, the budding yeast ATR orthologue Mec1 confers resistance to proteotoxic stress, suggesting an evolutionarily conserved role of the DNA damage response signaling pathway in proteostasis [313].
Notably, several proteins that form pathological aggregates have functions in DNA metabolism and genome maintenance. It is believed that cytosolic aggregation of these proteins may impair their nuclear functions in preserving genomic integrity of neurons, leading to build-up of DNA damage and induction of regulated cell death. For example, TDP-43, which exhibits nuclear exit and cytosolic aggregation in motor neurons in tau-negative FTLD patients and in 95% of ALS patients [314], is involved in DSB repair by NHEJ [305]. Loss of TDP-43 nuclear localization results in persistent DNA damage and prolonged activation of DNA damage response signaling, ultimately leading to neuronal death [305]. FUS, whose defects are associated with ~ 5% of familial ALS and ~ 1% of sporadic ALS cases due to loss of its nuclear localization and subsequent cytosolic aggregation, has been shown to facilitate DNA ligation during the final step of oxidative damage repair [315] and to be required for DSB repair by NHEJ and HR [316, 317]. HD-associated wild-type HTT is recruited to sites of DNA damage, colocalizes with BER proteins during oxidative stress [318], and serves as a scaffold protein for the transcription-coupled DNA repair (TCR) complex [319]. αSyn forms nuclear foci, which colocalize with γH2AX, and enhances ligation of DNA ends in vitro, and repair of bleomycin-induced DSBs is compromised in human HAP1 cells with αSyn knock-out [320]. Tau is involved in the organization of the microtubule cytoskeleton and the regulation of protein trafficking on microtubules [321], and may have a general role in microtubule-mediated transport of DNA repair proteins into the nucleus, as shown for the 53BP1 protein required for NHEJ [160, 322]. Tau is also implicated in the protection of neuronal DNA by an as yet poorly understood mechanism [323].
At the same time, protein aggregates may directly or indirectly cause DNA damage. Using in vitro models of PD, it was shown that αSyn overexpression induces DNA breaks in the presence of pro-oxidant Fe salts and that the DNA nicking property of αSyn is enhanced by its misfolding [324]. Pre-formed fibrils (PFF) of αSyn activates NOS, leading to oxidative DNA damage and PARP-1 activation. Excessive PAR levels, as well as PARylation of αSyn PFF, not only accelerates αSyn aggregation but also leads to neuronal cell death via parthanatos [325]. Interestingly, lack of ATM in concert with elevated ROS triggers the accumulation of transcription-dependent SSBs followed by increased protein PARylation at sites of DNA damage and subsequent protein aggregation [311]. Consequently, PARP-1 inhibition shows neuroprotective effects not only in PD [325] but also in ALS [326] and HD [327]. A recent study showed that αSyn aggregation probably induces DSBs, as indicated by increased formation of γH2AX foci, both in mixed glial cultures and in the αSyn PFF mouse model of PD. This is followed by activation of the cyclic GMP–AMP synthase (cGAS)/stimulator of interferon genes (STING), resulting in neuroinflammation that is known to contribute to neurodegeneration in PD [302]. Protein aggregation may also negatively affect DNA repair pathways indirectly by decreasing the levels of DDR proteins or by sequestering DDR proteins in the cytoplasm of neurons. For example, BRCA1, a key factor in DSB repair by HR, is sequestered into cytosolic inclusions of hyperphoshorylated tau in AD brain samples [328], while reduced BRCA1 levels are observed in neurons exposed to Aβ oligomers and in AD patients [303].
Another source of protein aggregate-derived DNA damage could be ROS. It is well established that mitochondrial dysfunction and increased oxidative stress contribute to neurodegenerative disorders and aging of the human brain [329,330,331]. Elevated oxidative damage to mitochondrial and nuclear DNA was observed in brain tissue from AD, PD, HD, and ALS patients and rodent models [309, 332, 333]. The origin of increased ROS production and the mechanisms underlying redox imbalances remain elusive, but most likely involve physical interactions of protein aggregates with mitochondria, leading to impaired mitochondrial metabolism [309, 330, 331, 333], and/or aggregation of mutant superoxide dismutase SOD1, as observed in some ALS cases, resulting in nuclear depletion of wild type SOD1 [334].
Taken together, accumulating lines of evidence suggest that genome instability and proteome instability are interrelated and jointly contribute to neurodegeneration and carcinogenesis (Fig. 3). Since cells exposed to As(III) and Sb(III) suffer from both genotoxic and proteotoxic damage, the possibility that both modes of toxic action are mechanistically related and together contribute to neurotoxicity and the development of neurodegenerative disorders should be considered. Importantly, the negative effects of metalloids on cells, including neurons, largely overlap with hallmarks of neurodegeneration. The genotoxic properties of metalloids, such as oxidative stress-derived DNA damage, inhibition of DNA repair mechanisms, induction of topoisomerase–DNA adducts, and perturbation of the cytoskeleton, may lead to genome instability and subsequent overload of PQC systems and increased protein aggregation, as observed under various genotoxic conditions [312]. At the same time, metalloids induce pathological protein aggregation as well as global protein misfolding and aggregation, which may also negatively impact the PQC systems and further promote genome and proteome instability. Thus, the genotoxic and proteotoxic effects of metalloids may act as a double-edged sword with significant potential to both initiate and accelerate neurodegeneration.
Conclusions and future perspectives
In this review, we highlighted how the metalloids arsenic and antimony cause genotoxicity and proteotoxicity and presented evidence that they may contribute to disease in a concerted way. Our understanding of how metalloids cause genotoxic and proteotoxic stress and the ensuing human health impacts, has increased considerably in the past decades. However, several key aspects of metalloid toxicity and cellular defence remain incompletely understood. For example, an important area for future studies is to clarify aggregate toxicity mechanisms, specifically the structures and properties of the protein aggregates formed during metalloid exposure that contribute to their toxicity as well as the features of these aggregates that limit aggregate binding and processing by chaperones and perhaps other PQC factors. A second key question concerns sensing and signaling mechanisms, specifically how cells detect metalloids and/or metalloid-induced cellular damage and how they couple the sensing mechanisms to the regulation of protective systems. The third question concerns the generality of the observations that metals and metalloids can perturb protein folding in cells, specifically which metals affect proteostasis in vivo and the mechanisms whereby the cause proteotoxicity. Loss of proteome and genome integrity are common hallmarks of neurodegenerative diseases, aging, and cancer. Given that certain metals and metalloids can cause proteotoxic and genotoxic effects, suggest that metals may be major environmental factors driving these diseases. Thus, future efforts should be directed toward elucidating mechanisms by which metalloids initiate or accelerate neurodegeneration and other human diseases. Similarly, how metal/metalloid-induced genotoxic and proteotoxic stress are interrelated and jointly contribute to neurodegeneration and carcinogenesis remains to be clarified. Finally, epidemiological studies are needed to cement the importance of individual metals as risk factors for disease, and regulatory measures should be taken to reduce exposure levels. Molecular insights into cellular toxicity and response mechanisms and their links to pathogenesis especially at low environmentally-relevant concentrations, may pave the way for the development of strategies for both disease prevention and treatment.
Data availability
Not applicable.
Abbreviations
- 8-OHdG:
-
8-Hydroxyl-2′-deoxyguanine
- Aβ:
-
Amyloid β
- ABC:
-
ATP-binding cassette
- AD:
-
Alzheimer’s disease
- AIRAP:
-
Arsenite-inducible RNA-associated protein
- αSyn:
-
α-Synuclein
- ALS:
-
Amyotrophic lateral sclerosis
- APL:
-
Acute promyelocytic leukemia
- APP:
-
Amyloid precursor protein
- As(V):
-
Arsenate
- As(III):
-
Arsenite
- ATP:
-
Adenosine triphosphate
- BER:
-
Base excision repair
- cGAS:
-
Cyclic GMP–AMP synthase
- DDR:
-
DNA damage response
- DDT:
-
DNA damage tolerance
- DMAs(III):
-
Dimethylarsonous acid
- DSB:
-
Double-strand break
- FET:
-
FUS, EWS, TAF15
- FTLD:
-
Frontotemporal lobar degeneration
- FUS:
-
Fused in sarcoma
- GSH:
-
Reduced glutathione
- GSSG:
-
Oxidized glutathione
- GTP:
-
Guanosine triphosphate
- HD:
-
Huntington disease
- HR:
-
Homologous recombination
- HRI:
-
Heme-regulated inhibitor kinase
- hTERT:
-
Human telomerase reverse transcriptase
- Htt:
-
Huntingtin
- ICR:
-
Interstrand crosslink repair
- IPOD:
-
Insoluble protein deposit
- JUNQ:
-
Juxtanuclear protein quality control
- LRRK2:
-
Leucine-rich repeat kinase 2
- MMAs(III):
-
Monomethylarsonous acid
- MMR:
-
Mismatch repair
- NER:
-
Nucleotide excision repair
- NHEJ:
-
Non-homologous end joining
- NOS:
-
Nitric oxide synthase
- NOX:
-
NADPH oxidase
- PAR:
-
Poly(ADP-ribose)
- PARP-1:
-
Poly(ADP-ribose) polymerase-1
- PB:
-
Processing body
- PD:
-
Parkinson’s disease
- PFF:
-
Pre-formed fibrils
- PML-NBs:
-
Promyelocytic leukemia nuclear bodies
- PQC:
-
Protein quality control
- RING:
-
Really interesting new gene
- RNS:
-
Reactive nitrogen species
- ROS:
-
Reactive oxygen species
- SAC:
-
Spindle assembly checkpoint
- Sb(III):
-
Antimonite
- SG:
-
Stress granule
- SSB:
-
Single-strand break
- STING:
-
Stimulator of interferon genes
- TCR:
-
Transcription-coupled DNA repair
- TDP-43:
-
TAR DNA binding protein 43
- TLS:
-
Translesion synthesis
- Top1:
-
DNA topoisomerase I
- TOP1cc:
-
DNA topoisomerase 1 cleavage complex
- TORC1:
-
TOR complex 1
- UPS:
-
Ubiquitin–proteasome system
- UVC:
-
Ultraviolet C radiation
- WHO:
-
World Health Organization
- XPA:
-
Xeroderma pigmentosum complementation group A protein
- ZnF:
-
Zinc finger
References
Oremland RS, Stolz JF (2003) The ecology of arsenic. Science 300:939–944
Pierart A, Shahid M, Sejalon-Delmas N, Dumat C (2015) Antimony bioavailability: knowledge and research perspectives for sustainable agricultures. J Hazard Mater 289:219–234
Chen QY, Costa M (2021) Arsenic: a global environmental challenge. Annu Rev Pharmacol 61:47–63
Shankar S, Shanker U, Shikha (2014) Arsenic contamination of groundwater: a review of sources, prevalence, health risks, and strategies for mitigation. Sci World J 2014:304524
Shaji E, Santosh M, Sarath KV, Prakash P, Deepchand V, Divya BV (2021) Arsenic contamination of groundwater: a global synopsis with focus on the Indian Peninsula. Geosci Front 12:101079
He MC, Wang XQ, Wu FC, Fu ZY (2012) Antimony pollution in China. Sci Tot Environ 421:41–50
Herath I, Vithanage M, Bundschuh J (2017) Antimony as a global dilemma: geochemistry, mobility, fate and transport. Environ Pollut 223:545–559
Podgorski J, Berg M (2020) Global threat of arsenic in groundwater. Science 368:845–850
Fowler BA, Chou C-HSJ, Jones RL, Costa M, Chen C-J (2022) Arsenic. In: Nordberg GF, Costa M (eds) Handbook on the toxicology of metals. Academic Press, Cambridge, pp 41–89
Khairul I, Wang QQ, Jiang YH, Wang C, Naranmandura H (2017) Metabolism, toxicity and anticancer activities of arsenic compounds. Oncotarget 8:23905–23926
Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano JH, Thompson C, Suk WA (2013) The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect 121:295–302
Chin-Chan M, Navarro-Yepes J, Quintanilla-Vega B (2015) Environmental pollutants as risk factors for neurodegenerative disorders: Alzheimer and Parkinson diseases. Front Cell Neurosci 9:124
Rahman MA, Hannan MA, Uddin MJ, Rahman MS, Rashid MM, Kim B (2021) Exposure to environmental arsenic and emerging risk of Alzheimer’s disease: perspective mechanisms, management strategy, and future directions. Toxics 9:188
Sundar S, Chakravarty J (2010) Antimony toxicity. Int J Environ Res Public Health 7:4267–4277
Tamás MJ (2016) Cellular and molecular mechanisms of antimony transport, toxicity and resistance. Environ Chem 13:955–962
Tylenda CA, Tomei Torres FA, Sullivan DWJ (2022) Antimony. In: Nordberg GF, Costa M (eds) Handbook on the toxicology of metals. Academic Press, Cambridge, pp 23–40
Zhou X, Speer RM, Volk L, Hudson LG, Liu KJ (2021) Arsenic co-carcinogenesis: inhibition of DNA repair and interaction with zinc finger proteins. Semin Cancer Biol 76:86–98
Pi JB, Kumagai Y, Sun GF, Yamauchi H, Yoshida T, Iso H, Endo A, Yu LY, Yuki KC, Miyauchi T, Shimojo N (2000) Decreased serum concentrations of nitric oxide metabolites among Chinese in an endemic area of chronic arsenic poisoning in Inner Mongolia. Free Radical Biol Med 28:1137–1142
Wu MM, Chiou HY, Wang TW, Hsueh YM, Wang IH, Chen CJ, Lee TC (2001) Association of blood arsenic levels with increased reactive oxidants and decreased antioxidant capacity in a human population of northeastern Taiwan. Environ Health Persp 109:1011–1017
Takayama Y, Masuzaki Y, Mizutani F, Iwata T, Maeda E, Tsukada M, Nomura K, Ito Y, Chisaki Y, Murata K (2021) Associations between blood arsenic and urinary arsenic species concentrations as an exposure characterization tool. Sci Total Environ 750:141517
Green BB, Karagas MR, Punshon T, Jackson BP, Robbins DJ, Houseman EA, Marsit CJ (2016) Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the New Hampshire birth cohort study (USA). Environ Health Persp 124:1253–1260
Shen ZX, Chen GQ, Ni JH, Li XS, Xiong SM, Qiu QY, Zhu J, Tang W, Sun GL, Yang KQ, Chen Y, Zhou L, Fang ZW, Wang YT, Ma J, Zhang P, Zhang TD, Chen SJ, Chen Z, Wang ZY (1997) Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL): II. Clinical efficacy and pharmacokinetics in relapsed patients. Blood 89:3354–3360
Neves DBD, Caldas ED, Sampaio RNR (2009) Antimony in plasma and skin of patients with cutaneous leishmaniasis—relationship with side effects after treatment with meglumine antimoniate. Trop Med Int Health 14:1515–1522
Fairlamb AH, Horn D (2018) Melarsoprol resistance in African trypanosomiasis. Trends Parasitol 34:481–492
Frezard F, Demicheli C, Kato KC, Reis PG, Lizarazo-Jaimes EH (2013) Chemistry of antimony-based drugs in biological systems and studies of their mechanism of action. Rev Inorg Chem 33:1–12
Paul NP, Galvan AE, Yoshinaga-Sakurai K, Rosen BP, Yoshinaga M (2023) Arsenic in medicine: past, present and future. Biometals 36:283–301
Fang Y, Zhang Z (2020) Arsenic trioxide as a novel anti-glioma drug: a review. Cell Mol Biol Lett 25:44
Jiang Y, Shen X, Zhi F, Wen Z, Gao Y, Xu J, Yang B, Bai Y (2023) An overview of arsenic trioxide-involved combined treatment algorithms for leukemia: basic concepts and clinical implications. Cell Death Discov 9:266
Hadjikakou SK, Ozturk II, Banti CN, Kourkoumelis N, Hadjiliadis N (2015) Recent advances on antimony(III/V) compounds with potential activity against tumor cells. J Inorg Biochem 153:293–305
Sharma P, Perez D, Cabrera A, Rosas N, Arias JL (2008) Perspectives of antimony compounds in oncology. Acta Pharmacol Sin 29:881–890
Lemire JA, Harrison JJ, Turner RJ (2013) Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol 11:371–384
Tamás MJ, Sharma KS, Ibstedt S, Jacobson T, Christen P (2014) Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 4:252–267
Shen S, Li XF, Cullen WR, Weinfeld M, Le XC (2013) Arsenic binding to proteins. Chem Rev 113:7769–7792
Chen QY, DesMarais T, Costa M (2019) Metals and mechanisms of carcinogenesis. Annu Rev Pharmacol Toxicol 59:537–554
Vergara-Geronimo CA, Leon Del Rio A, Rodriguez-Dorantes M, Ostrosky-Wegman P, Salazar AM (2021) Arsenic-protein interactions as a mechanism of arsenic toxicity. Toxicol Appl Pharmacol 431:115738
Kitchin KT, Wallace K (2008) The role of protein binding of trivalent arsenicals in arsenic carcinogenesis and toxicity. J Inorg Biochem 102:532–539
Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, Liang WX, Song AX, Lallemand-Breitenbach V, Jeanne M, Zhang QY, Yang HY, Huang QH, Zhou GB, Tong JH, Zhang Y, Wu JH, Hu HY, de The H, Chen SJ, Chen Z (2010) Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science 328:240–243
Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H (2008) Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 10:547–555
Kumar NV, Yang J, Pillai JK, Rawat S, Solano C, Kumar A, Grotli M, Stemmler TL, Rosen BP, Tamás MJ (2016) Arsenic directly binds to and activates the yeast AP-1-like transcription factor Yap8. Mol Cell Biol 36:913–922
Shi W, Wu J, Rosen BP (1994) Identification of a putative metal binding site in a new family of metalloregulatory proteins. J Biol Chem 269:19826–19829
Guerra-Moreno A, Prado MA, Ang J, Schnell HM, Micoogullari Y, Paulo JA, Finley D, Gygi SP, Hanna J (2019) Thiol-based direct threat sensing by the stress-activated protein kinase Hog1. Sci Signal. 12:eaaw4956
Yan S, Jin L, Sun H (2005) Antimony in medicine. In: Gielen M, Tiekink ER (eds) Metallotherapeutic drugs and metal-based diagnostic agents: the use of metals in medicine. Wiley, Chichester, pp 441–462
Beyersmann D, Hartwig A (2008) Carcinogenic metal compounds: recent insight into molecular and cellular mechanisms. Arch Toxicol 82:493–512
Boreiko CJ, Rossman TG (2020) Antimony and its compounds: health impacts related to pulmonary toxicity, cancer, and genotoxicity. Toxicol Appl Pharmacol 403:115156
Viswanathan T, Chen J, Wu M, An L, Kandavelu P, Sankaran B, Radhakrishnan M, Li M, Rosen BP (2021) Functional and structural characterization of AntR, an Sb(III) responsive transcriptional repressor. Mol Microbiol 116:427–437
Wysocki R, Fortier PK, Maciaszczyk E, Thorsen M, Leduc A, Odhagen A, Owsianik G, Ulaszewski S, Ramotar D, Tamás MJ (2004) Transcriptional activation of metalloid tolerance genes in Saccharomyces cerevisiae requires the AP-1-like proteins Yap1p and Yap8p. Mol Biol Cell 15:2049–2060
Bentley R, Chasteen TG (2002) Microbial methylation of metalloids: arsenic, antimony, and bismuth. Microbiol Mol Biol Rev 66:250–271
Thomas DJ (2021) Arsenic methylation—lessons from three decades of research. Toxicology 457:152800
Marapakala K, Qin J, Rosen BP (2012) Identification of catalytic residues in the As(III) S-adenosylmethionine methyltransferase. Biochemistry 51:944–951
Lee J, Levin DE (2018) Intracellular mechanism by which arsenite activates the yeast stress MAPK Hog1. Mol Biol Cell 29:1904–1915
Lee J, Levin DE (2022) Differential metabolism of arsenicals regulates Fps1-mediated arsenite transport. J Cell Biol 221:e202109034
Martinez VD, Vucic EA, Becker-Santos DD, Gil L, Lam WL (2011) Arsenic exposure and the induction of human cancers. J Toxicol 2011:431287
IARC Working Group on the Evaluation of Carcinogenic Risks to Humans (2012) Volume 100: Arsenic, metals, fibres, and dusts. International Agency for Research on Cancer, Lyon
Benbrahim-Tallaa L, Waalkes MP (2008) Inorganic arsenic and human prostate cancer. Environ Health Perspect 116:158–164
Liu J, Waalkes MP (2008) Liver is a target of arsenic carcinogenesis. Toxicol Sci 105:24–32
Saint-Jacques N, Parker L, Brown P, Dummer TJ (2014) Arsenic in drinking water and urinary tract cancers: a systematic review of 30 years of epidemiological evidence. Environ Health 13:44
Klein CB, Leszczynska J, Hickey C, Rossman TG (2007) Further evidence against a direct genotoxic mode of action for arsenic-induced cancer. Toxicol Appl Pharmacol 222:289–297
Tokar EJ, Benbrahim-Tallaa L, Ward JM, Lunn R, Sams RL 2nd, Waalkes MP (2010) Cancer in experimental animals exposed to arsenic and arsenic compounds. Crit Rev Toxicol 40:912–927
Tokar EJ, Diwan BA, Ward JM, Delker DA, Waalkes MP (2011) Carcinogenic effects of “whole-life” exposure to inorganic arsenic in CD1 mice. Toxicol Sci 119:73–83
Waalkes MP, Qu W, Tokar EJ, Kissling GE, Dixon D (2014) Lung tumors in mice induced by “whole-life” inorganic arsenic exposure at human-relevant doses. Arch Toxicol 88:1619–1629
Rossman TG (2003) Mechanism of arsenic carcinogenesis: an integrated approach. Mutat Res 533:37–65
Litwin I, Bocer T, Dziadkowiec D, Wysocki R (2013) Oxidative stress and replication-independent DNA breakage induced by arsenic in Saccharomyces cerevisiae. PLoS Genet 9:e1003640
Rossman TG, Klein CB (2011) Genetic and epigenetic effects of environmental arsenicals. Metallomics 3:1135–1141
Ozturk M, Metin M, Altay V, Bhat RA, Ejaz M, Gul A, Unal BT, Hasanuzzaman M, Nibir L, Nahar K, Bukhari A, Dervash MA, Kawano T (2022) Arsenic and human health: genotoxicity, epigenomic effects, and cancer signaling. Biol Trace Elem Res 200:988–1001
Saerens A, Ghosh M, Verdonck J, Godderis L (2019) Risk of cancer for workers exposed to antimony compounds: a systematic review. Int J Environ Res Public Health 16:4474
National Toxicology Program (NTP) (2018) Report on carcinogens monograph on antimony trioxide. In: Program NT (ed) RoC monograph series. National Toxicology Program, Research Triangle Park
IARC Monographs on the Identification of Carcinogenic Hazards to Humans (2023) Volume 131: cobalt, antimony compounds, and weapons-grade tungsten alloy. International Agency for Research on Cancer, Lyon
Litwin I, Mucha S, Pilarczyk E, Wysocki R, Maciaszczyk-Dziubinska E (2021) Complex mechanisms of antimony genotoxicity in budding yeast involves replication and topoisomerase I-associated DNA lesions, telomere dysfunction and inhibition of DNA repair. Int J Mol Sci 22:4510
Flora SJ (2011) Arsenic-induced oxidative stress and its reversibility. Free Radic Biol Med 51:257–281
Hu Y, Li J, Lou B, Wu R, Wang G, Lu C, Wang H, Pi J, Xu Y (2020) The role of reactive oxygen species in arsenic toxicity. Biomolecules 10:240
Li X, Pi J, Li B, Xu Y, Jin Y, Sun G (2008) Urinary arsenic speciation and its correlation with 8-OHdG in Chinese residents exposed to arsenic through coal burning. Bull Environ Contamin Toxicol 81:406–411
Ruiz-Ramos R, Lopez-Carrillo L, Rios-Perez AD, De Vizcaya-Ruiz A, Cebrian ME (2009) Sodium arsenite induces ROS generation, DNA oxidative damage, HO-1 and c-Myc proteins, NF-kappaB activation and cell proliferation in human breast cancer MCF-7 cells. Mutat Res 674:109–115
Dodson M, de la Vega MR, Harder B, Castro-Portuguez R, Rodrigues SD, Wong PK, Chapman E, Zhang DD (2018) Low-level arsenic causes proteotoxic stress and not oxidative stress. Toxicol Appl Pharmacol 341:106–113
Ying S, Myers K, Bottomley S, Helleday T, Bryant HE (2009) BRCA2-dependent homologous recombination is required for repair of Arsenite-induced replication lesions in mammalian cells. Nucleic Acids Res 37:5105–5113
Lynn S, Gurr JR, Lai HT, Jan KY (2000) NADH oxidase activation is involved in arsenite-induced oxidative DNA damage in human vascular smooth muscle cells. Circ Res 86:514–519
Chou WC, Jie C, Kenedy AA, Jones RJ, Trush MA, Dang CV (2004) Role of NADPH oxidase in arsenic-induced reactive oxygen species formation and cytotoxicity in myeloid leukemia cells. Proc Natl Acad Sci USA 101:4578–4583
Liu SX, Davidson MM, Tang X, Walker WF, Athar M, Ivanov V, Hei TK (2005) Mitochondrial damage mediates genotoxicity of arsenic in mammalian cells. Cancer Res 65:3236–3242
Ding W, Hudson LG, Liu KJ (2005) Inorganic arsenic compounds cause oxidative damage to DNA and protein by inducing ROS and RNS generation in human keratinocytes. Mol Cell Biochem 279:105–112
Ding W, Hudson LG, Sun X, Feng C, Liu KJ (2008) As(III) inhibits ultraviolet radiation-induced cyclobutane pyrimidine dimer repair via generation of nitric oxide in human keratinocytes. Free Radic Biol Med 45:1065–1072
Soest DMKv, Polderman PE, Toom WTFd, Zwakenberg S, Henau Sd, Burgering BMT, Dansen TB (2023) Mitochondrial H2O2 release does not directly cause genomic DNA damage, bioRxiv, 2023.03.29.534749
Yamanaka K, Okada S (1994) Induction of lung-specific DNA-damage by metabolically methylated arsenics via the production of free-radicals. Environ Health Persp 102:37–40
Yamanaka K, Hayashi H, Tachikawa M, Kato K, Hasegawa A, Oku N, Okada S (1997) Metabolic methylation is a possible genotoxicity-enhancing process of inorganic arsenics. Mutat Res-Gen Tox En 394:95–101
Cao D, Zhao J, Nguyan LN, Nguyen LNT, Khanal S, Dang X, Schank M, Chand Thakuri BK, Wu XY, Morrison ZD, El Gazzar M, Zou Y, Ning S, Wang L, Moorman JP, Yao ZQ (2019) Disruption of telomere integrity and DNA repair machineries by KML001 induces T cell senescence, apoptosis, and cellular dysfunctions. Front Immunol 10:1152
Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, Agama K, Nitiss JL, Redon C (2006) Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol 81:179–229
Wai KM, Swe T, Myar MT, Aisyah CR, Hninn TSS (2022) Telomeres susceptibility to environmental arsenic exposure: shortening or lengthening? Front Public Health 10:1059248
Cheng Y, Li Y, Ma C, Song Y, Xu H, Yu H, Xu S, Mu Q, Li H, Chen Y, Zhao G (2016) Arsenic trioxide inhibits glioma cell growth through induction of telomerase displacement and telomere dysfunction. Oncotarget 7:12682–12692
Liu L, Trimarchi JR, Navarro P, Blasco MA, Keefe DL (2003) Oxidative stress contributes to arsenic-induced telomere attrition, chromosome instability, and apoptosis. J Biol Chem 278:31998–32004
Phatak P, Dai F, Butler M, Nandakumar MP, Gutierrez PL, Edelman MJ, Hendriks H, Burger AM (2008) KML001 cytotoxic activity is associated with its binding to telomeric sequences and telomere erosion in prostate cancer cells. Clin Cancer Res 14:4593–4602
Jeon BG, Kumar BM, Kang EJ, Maeng GH, Lee YM, Hah YS, Ock SA, Kwack DO, Park BW, Rho GJ (2011) Differential cytotoxic effects of sodium meta-arsenite on human cancer cells, dental papilla stem cells and somatic cells correlate with telomeric properties and gene expression. Anticancer Res 31:4315–4328
Kuroda K, Endo G, Okamoto A, Yoo YS, Horiguchi S (1991) Genotoxicity of beryllium, gallium and antimony in short-term assays. Mutat Res 264:163–170
Koch B, Maser E, Hartwig A (2017) Low concentrations of antimony impair DNA damage signaling and the repair of radiation-induced DSB in HeLa S3 cells. Arch Toxicol 91:3823–3833
Kopp B, Zalko D, Audebert M (2018) Genotoxicity of 11 heavy metals detected as food contaminants in two human cell lines. Environ Mol Mutagen 59:202–210
Boreiko CJ, Hendriks G, Derr R, Huppert M, Rossman TG (2021) Mode of action assessment of the genotoxic properties of antimony and its compounds evaluated in the ToxTracker assay. Mutat Res Genet Toxicol Environ Mutagen 865:503333
Wyllie S, Fairlamb AH (2006) Differential toxicity of antimonial compounds and their effects on glutathione homeostasis in a human leukaemia monocyte cell line. Biochem Pharmacol 71:257–267
Hashemzaei M, Pourahmad J, Safaeinejad F, Tabrizian K, Akbari F, Bagheri G, Hosseini MJ, Shahraki J (2015) Antimony induces oxidative stress and cell death in normal hepatocytes. Toxicol Environ Chem 97:256–265
Yao Q, Yang A, Hu X, Zou H, Chen J, Li Q, Lv S, Yu X, Li C (2023) Effects of antimony exposure on DNA damage and genome-wide variation in zebrafish (Danio rerio) liver. Aquat Toxicol 259:106524
Pizzul P, Casari E, Gnugnoli M, Rinaldi C, Corallo F, Longhese MP (2022) The DNA damage checkpoint: a tale from budding yeast. Front Genet 13:995163
Eki T (2018) Yeast-based genotoxicity tests for assessing DNA alterations and DNA stress responses: a 40-year overview. Appl Microbiol Biotechnol 102:2493–2507
Vanderwaeren L, Dok R, Voordeckers K, Nuyts S, Verstrepen KJ (2022) Saccharomyces cerevisiae as a model system for eukaryotic cell biology, from cell cycle control to DNA damage response. Int J Mol Sci 23:11665
Sordet O, Liao Z, Liu H, Antony S, Stevens EV, Kohlhagen G, Fu H, Pommier Y (2004) Topoisomerase I-DNA complexes contribute to arsenic trioxide-induced apoptosis. J Biol Chem 279:33968–33975
Maciaszczyk-Dziubinska E, Wawrzycka D, Wysocki R (2012) Arsenic and antimony transporters in eukaryotes. Int J Mol Sci 13:3527–3548
Lee TC, Ho IC (1994) Differential cytotoxic effects of arsenic on human and animal-cells. Environ Health Persp 102:101–105
Drobna Z, Walton FS, Harmon AW, Thomas DJ, Styblo M (2010) Interspecies differences in metabolism of arsenic by cultured primary hepatocytes. Toxicol Appl Pharm 245:47–56
Thomas DJ, Li JX, Waters SB, Xing WB, Adair BM, Drobna Z, Devesa V, Styblo M (2007) Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp Biol Med 232:3–13
Tam LM, Price NE, Wang Y (2020) Molecular mechanisms of arsenic-induced disruption of DNA repair. Chem Res Toxicol 33:709–726
Selmin OI, Donovan MG, Skovan B, Paine-Murieta GD, Romagnolo DF (2019) Arsenic-induced BRCA1 CpG promoter methylation is associated with the downregulation of ERalpha and resistance to tamoxifen in MCF7 breast cancer cells and mouse mammary tumor xenografts. Int J Oncol 54:869–878
Ding X, Zhang A, Li C, Ma L, Tang S, Wang Q, Yang G, Li J (2021) The role of H3K9me2-regulated base excision repair genes in the repair of DNA damage induced by arsenic in HaCaT cells and the effects of Ginkgo biloba extract intervention. Environ Toxicol 36:850–860
Mauro M, Caradonna F, Klein CB (2016) Dysregulation of DNA methylation induced by past arsenic treatment causes persistent genomic instability in mammalian cells. Environ Mol Mutagen 57:137–150
Bhattacharjee P, Sanyal T, Bhattacharjee S, Bhattacharjee P (2018) Epigenetic alteration of mismatch repair genes in the population chronically exposed to arsenic in West Bengal India. Environ Res 163:289–296
Vilas CK, Emery LE, Denchi EL, Miller KM (2018) Caught with one’s zinc fingers in the genome integrity cookie jar. Trends Genet 34:313–325
Zhou X, Sun X, Cooper KL, Wang F, Liu KJ, Hudson LG (2011) Arsenite interacts selectively with zinc finger proteins containing C3H1 or C4 motifs. J Biol Chem 286:22855–22863
Zhou X, Sun X, Mobarak C, Gandolfi AJ, Burchiel SW, Hudson LG, Liu KJ (2014) Differential binding of monomethylarsonous acid compared to arsenite and arsenic trioxide with zinc finger peptides and proteins. Chem Res Toxicol 27:690–698
Sun X, Zhou X, Du L, Liu W, Liu Y, Hudson LG, Liu KJ (2014) Arsenite binding-induced zinc loss from PARP-1 is equivalent to zinc deficiency in reducing PARP-1 activity, leading to inhibition of DNA repair. Toxicol Appl Pharmacol 274:313–318
Walter I, Schwerdtle T, Thuy C, Parsons JL, Dianov GL, Hartwig A (2007) Impact of arsenite and its methylated metabolites on PARP-1 activity, PARP-1 gene expression and poly(ADP-ribosyl)ation in cultured human cells. DNA Repair (Amst) 6:61–70
Qin XJ, Hudson LG, Liu W, Timmins GS, Liu KJ (2008) Low concentration of arsenite exacerbates UVR-induced DNA strand breaks by inhibiting PARP-1 activity. Toxicol Appl Pharmacol 232:41–50
Ding W, Liu W, Cooper KL, Qin XJ, de Souza Bergo PL, Hudson LG, Liu KJ (2009) Inhibition of poly(ADP-ribose) polymerase-1 by arsenite interferes with repair of oxidative DNA damage. J Biol Chem 284:6809–6817
Wang F, Zhou X, Liu W, Sun X, Chen C, Hudson LG, Jian Liu K (2013) Arsenite-induced ROS/RNS generation causes zinc loss and inhibits the activity of poly(ADP-ribose) polymerase-1. Free Radic Biol Med 61:249–256
Zhou X, Cooper KL, Sun X, Liu KJ, Hudson LG (2015) Selective sensitization of zinc finger protein oxidation by reactive oxygen species through arsenic binding. J Biol Chem 290:18361–18369
Zhou X, Cooper KL, Huestis J, Xu H, Burchiel SW, Hudson LG, Liu KJ (2016) S-nitrosation on zinc finger motif of PARP-1 as a mechanism of DNA repair inhibition by arsenite. Oncotarget 7:80482–80492
Zhou X, Ding X, Shen J, Yang D, Hudson LG, Liu KJ (2019) Peroxynitrite contributes to arsenic-induced PARP-1 inhibition through ROS/RNS generation. Toxicol Appl Pharmacol 378:114602
Ray Chaudhuri A, Nussenzweig A (2017) The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 18:610–621
Ding X, Zhou X, Cooper KL, Huestis J, Hudson LG, Liu KJ (2017) Differential sensitivities of cellular XPA and PARP-1 to arsenite inhibition and zinc rescue. Toxicol Appl Pharmacol 331:108–115
Roy JS, Chatterjee D, Das N, Giri AK (2018) Substantial evidences indicate that inorganic arsenic is a genotoxic carcinogen: a review. Toxicol Res 34:311–324
Poonepalli A, Balakrishnan L, Khaw AK, Low GK, Jayapal M, Bhattacharjee RN, Akira S, Balajee AS, Hande MP (2005) Lack of poly(ADP-ribose) polymerase-1 gene product enhances cellular sensitivity to arsenite. Cancer Res 65:10977–10983
Borszekova Pulzova L, Ward TA, Chovanec M (2020) XPA: DNA repair protein of significant clinical importance. Int J Mol Sci 21:2182
Asmuss M, Mullenders LH, Eker A, Hartwig A (2000) Differential effects of toxic metal compounds on the activities of Fpg and XPA, two zinc finger proteins involved in DNA repair. Carcinogenesis 21:2097–2104
Mustra DJ, Warren AJ, Wilcox DE, Hamilton JW (2007) Preferential binding of human XPA to the mitomycin C-DNA interstrand crosslink and modulation by arsenic and cadmium. Chem-Biol Interact 168:159–168
Piatek K, Schwerdtle T, Hartwig A, Bal W (2008) Monomethylarsonous acid destroys a tetrathiolate zinc finger much more efficiently than inorganic arsenite: mechanistic considerations and consequences for DNA repair inhibition. Chem Res Toxicol 21:600–606
Huestis J, Zhou X, Chen L, Feng C, Hudson LG, Liu KJ (2016) Kinetics and thermodynamics of zinc(II) and arsenic(III) binding to XPA and PARP-1 zinc finger peptides. J Inorg Biochem 163:45–52
Zhang F, Paramasivam M, Cai Q, Dai X, Wang P, Lin K, Song J, Seidman MM, Wang Y (2014) Arsenite binds to the RING finger domains of RNF20-RNF40 histone E3 ubiquitin ligase and inhibits DNA double-strand break repair. J Am Chem Soc 136:12884–12887
Jiang J, Bellani M, Li L, Wang P, Seidman MM, Wang Y (2017) Arsenite binds to the RING finger domain of FANCL E3 ubiquitin ligase and inhibits DNA interstrand crosslink repair. ACS Chem Biol 12:1858–1866
Volk LB, Cooper KL, Jiang T, Paffett ML, Hudson LG (2022) Impacts of arsenic on Rad18 and translesion synthesis. Toxicol Appl Pharmacol 454:116230
Tam LM, Jiang J, Wang P, Li L, Miao W, Dong X, Wang Y (2017) Arsenite binds to the zinc finger motif of TIP60 histone acetyltransferase and induces its degradation via the 26S proteasome. Chem Res Toxicol 30:1685–1693
Rossetto D, Truman AW, Kron SJ, Cote J (2010) Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin Cancer Res 16:4543–4552
Grosskopf C, Schwerdtle T, Mullenders LH, Hartwig A (2010) Antimony impairs nucleotide excision repair: XPA and XPE as potential molecular targets. Chem Res Toxicol 23:1175–1183
Demicheli C, Frezard F, Mangrum JB, Farrell NP (2008) Interaction of trivalent antimony with a CCHC zinc finger domain: potential relevance to the mechanism of action of antimonial drugs. Chem Commun 39:4828–4830
Frezard F, Silva H, Pimenta AM, Farrell N, Demicheli C (2012) Greater binding affinity of trivalent antimony to a CCCH zinc finger domain compared to a CCHC domain of kinetoplastid proteins. Metallomics 4:433–440
Müller S, Miller WH Jr, Dejean A (1998) Trivalent antimonials induce degradation of the PML-RAR oncoprotein and reorganization of the promyelocytic leukemia nuclear bodies in acute promyelocytic leukemia NB4 cells. Blood 92:4308–4316
Yang C, Hao R, Lan YF, Chen YJ, Wang C, Bu N, Wang QQ, Hussain L, Ma L, Maimaitiyiming Y, Lu XY, Naranmandura H (2019) Integrity of zinc finger motifs in PML protein is necessary for inducing its degradation by antimony. Metallomics 11:1419–1429
Takahashi S, Sato H, Kubota Y, Utsumi H, Bedford JS, Okayasu R (2002) Inhibition of DNA-double strand break repair by antimony compounds. Toxicology 180:249–256
States JC (2015) Disruption of mitotic progression by arsenic. Biol Trace Elem Res 166:34–40
Wise SS, Wise JP (2010) Aneuploidy as an early mechanistic event in metal carcinogenesis. Biochem Soc Trans 38:1650–1654
Li W, Chou IN (1992) Effects of sodium arsenite on the cytoskeleton and cellular glutathione levels in cultured cells. Toxicol Appl Pharmacol 114:132–139
Ramirez P, Eastmond DA, Laclette JP, Ostrosky-Wegman P (1997) Disruption of microtubule assembly and spindle formation as a mechanism for the induction of aneuploid cells by sodium arsenite and vanadium pentoxide. Mutat Res 386:291–298
Zhang X, Yang F, Shim JY, Kirk KL, Anderson DE, Chen X (2007) Identification of arsenic-binding proteins in human breast cancer cells. Cancer Lett 255:95–106
Kligerman AD, Doerr CL, Tennant AH (2005) Oxidation and methylation status determine the effects of arsenic on the mitotic apparatus. Mol Cell Biochem 279:113–121
Zhao YZ, Toselli P, Li WD (2012) Microtubules as a critical target for arsenic toxicity in lung cells in vitro and in vivo. Int J Environ Res Public Health 9:474–495
Yih LH, Ho IC, Lee TC (1997) Sodium arsenite disturbs mitosis and induces chromosome loss in human fibroblasts. Cancer Res 57:5051–5059
Taylor BF, McNeely SC, Miller HL, States JC (2008) Arsenite-induced mitotic death involves stress response and is independent of tubulin polymerization. Toxicol Appl Pharmacol 230:235–246
Thorsen M, Perrone GG, Kristiansson E, Traini M, Ye T, Dawes IW, Nerman O, Tamás MJ (2009) Genetic basis of arsenite and cadmium tolerance in Saccharomyces cerevisiae. BMC Genomics 10:105
Pan X, Reissman S, Douglas NR, Huang Z, Yuan DS, Wang X, McCaffery JM, Frydman J, Boeke JD (2010) Trivalent arsenic inhibits the functions of chaperonin complex. Genetics 186:725–734
Wu JG, Cardoso APF, States VAR, Al-Eryani L, Doll M, Wise SS, Rai SN, States JC (2019) Overexpression of hsa-miR-186 induces chromosomal instability in arsenic-exposed human keratinocytes. Toxicol Appl Pharm 378:114614
Cardoso APF, Nail AN, Banerjee M, Wise SS, States JC (2023) miR-186 induces tetraploidy in arsenic exposed human keratinocytes. Ecotox Environ Saf 256:114823
Menzel DB, Hamadeh HK, Lee E, Meacher DM, Said V, Rasmussen RE, Greene H, Roth RN (1999) Arsenic binding proteins from human lymphoblastoid cells. Toxicol Lett 105:89–101
Izdebska M, Grzanka D, Gackowska L, Zuryn A, Grzanka A (2009) The influence of Trisenox on actin organization in HL-60 cells. Cent Eur J Biol 4:351–361
Qian Y, Liu KJ, Chen Y, Flynn DC, Castranova V, Shi X (2005) Cdc42 regulates arsenic-induced NADPH oxidase activation and cell migration through actin filament reorganization. J Biol Chem 280:3875–3884
Farah ME, Sirotkin V, Haarer B, Kakhniashvili D, Amberg DC (2011) Diverse protective roles of the actin cytoskeleton during oxidative stress. Cytoskeleton 68:340–354
Balta E, Kramer J, Samstag Y (2021) Redox regulation of the actin cytoskeleton in cell migration and adhesion: on the way to a spatiotemporal view. Front Cell Dev Biol 8:618261
Caridi CP, Plessner M, Grosse R, Chiolo I (2019) Nuclear actin filaments in DNA repair dynamics. Nat Cell Biol 21:1068–1077
Poruchynsky MS, Komlodi-Pasztor E, Trostel S, Wilkerson J, Regairaz M, Pommier Y, Zhang X, Kumar Maity T, Robey R, Burotto M, Sackett D, Guha U, Fojo AT (2015) Microtubule-targeting agents augment the toxicity of DNA-damaging agents by disrupting intracellular trafficking of DNA repair proteins. Proc Natl Acad Sci U S A 112:1571–1576
Kim JM (2022) Molecular link between DNA damage response and microtubule dynamics. Int J Mol Sci 23:6986
Huang YL, Zhang JL, McHenry KT, Kim MM, Zeng WQ, Lopez-Pajares V, Dibble CC, Mizgerd JP, Yuan ZM (2008) Induction of cytoplasmic accumulation of p53: a mechanism for low levels of arsenic exposure to predispose cells for malignant transformation. Cancer Res 68:9131–9136
Hamadeh HK, Vargas M, Lee E, Menzel DB (1999) Arsenic disrupts cellular levels of p53 and mdm2: a potential mechanism of carcinogenesis. Biochem Biophys Res Commun 263:446–449
Ferrario D, Gribaldo L, Hartung T (2016) Arsenic exposure and immunotoxicity: a review including the possible influence of age and sex. Curr Environ Health Rep 3:1–12
Balchin D, Hayer-Hartl M, Hartl FU (2016) In vivo aspects of protein folding and quality control. Science 353:aac4354
Hipp MS, Kasturi P, Hartl FU (2019) The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol 20:421–435
Chiti F, Dobson CM (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem 86:27–68
Finka A, Mattoo RUH, Goloubinoff P (2016) Experimental milestones in the discovery of molecular chaperones as polypeptide unfolding enzymes. Ann Rev Biochem 85:715–742
Sontag EM, Samant RS, Frydman J (2017) Mechanisms and functions of spatial protein quality control. Annu Rev Biochem 86:97–122
Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426:895–899
Ramadan D, Cline DJ, Bai S, Thorpe C, Schneider JP (2007) Effects of As(III) binding on beta-hairpin structure. J Am Chem Soc 129:2981–2988
Cline DJ, Thorpe C, Schneider JP (2003) Effects of As(III) binding on alpha-helical structure. J Am Chem Soc 125:2923–2929
Ramadan D, Rancy PC, Nagarkar RP, Schneider JP, Thorpe C (2009) Arsenic(III) species inhibit oxidative protein folding in vitro. Biochemistry 48:424–432
Sapra A, Ramadan D, Thorpe C (2015) Multivalency in the inhibition of oxidative protein folding by arsenic(III) species. Biochemistry 54:612–621
Jacobson T, Navarrete C, Sharma SK, Sideri TC, Ibstedt S, Priya S, Grant CM, Christen P, Goloubinoff P, Tamás MJ (2012) Arsenite interferes with protein folding and triggers formation of protein aggregates in yeast. J Cell Sci 125:5073–5083
Hua S, Klosowska A, Rodrigues JI, Petelski G, Esquembre LA, Lorentzon E, Olsen LF, Liberek K, Tamás MJ (2022) Differential contributions of the proteasome, autophagy, and chaperones to the clearance of arsenite-induced protein aggregates in yeast. J Biol Chem 298:102680
Ibstedt S, Sideri TC, Grant CM, Tamás MJ (2014) Global analysis of protein aggregation in yeast during physiological conditions and arsenite stress. Biol Open 3:913–923
Andersson S, Romero A, Rodrigues JI, Hua S, Hao X, Jacobson T, Karl V, Becker N, Ashouri A, Rauch S, Nyström T, Liu B, Tamás MJ (2021) Genome-wide imaging screen uncovers molecular determinants of arsenite-induced protein aggregation and toxicity. J Cell Sci 134:jcs258338
Rodrigues JI, Lorentzon E, Hua S, Boucher A, Tamás MJ (2023) Yeast chaperones and ubiquitin ligases contribute to proteostasis during arsenite stress by preventing or clearing protein aggregates. FEBS Lett 595:1733–1747
Weids AJ, Ibstedt S, Tamás MJ, Grant CM (2016) Distinct stress conditions result in aggregation of proteins with similar properties. Sci Rep 6:24554
Zhang HN, Yang L, Ling JY, Czajkowsky DM, Wang JF, Zhang XW, Zhou YM, Ge F, Yang MK, Xiong Q, Guo SJ, Le HY, Wu SF, Yan W, Liu B, Zhu H, Chen Z, Tao SC (2015) Systematic identification of arsenic-binding proteins reveals that hexokinase-2 is inhibited by arsenic. Proc Natl Acad Sci U S A 112:15084–15089
Hanna J, Waterman D, Isasa M, Elsasser S, Shi Y, Gygi S, Finley D (2014) Cuz1/Ynl155w, a zinc-dependent ubiquitin-binding protein, protects cells from metalloid-induced proteotoxicity. J Biol Chem 289:1876–1885
Guerra-Moreno A, Isasa M, Bhanu MK, Waterman DP, Eapen VV, Gygi SP, Hanna J (2015) Proteomic analysis identifies ribosome reduction as an effective proteotoxic stress response. J Biol Chem 290:29695–29706
Sok J, Calfon M, Lu J, Lichtlen P, Clark SG, Ron D (2001) Arsenite-inducible RNA-associated protein (AIRAP) protects cells from arsenite toxicity. Cell Stress Chaperones 6:6–15
Stanhill A, Haynes CM, Zhang Y, Min G, Steele MC, Kalinina J, Martinez E, Pickart CM, Kong XP, Ron D (2006) An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol Cell 23:875–885
Kim MN, Choi J, Ryu HW, Ryu KY (2015) Disruption of polyubiquitin gene Ubc leads to attenuated resistance against arsenite-induced toxicity in mouse embryonic fibroblasts. Biochim Biophys Acta 1853:996–1009
McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ (2005) Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280:16925–16933
Lorentzon E, Horvath I, Kumar R, Rodrigues JI, Tamás MJ, Wittung-Stafshede P (2021) Effects of the toxic metals arsenite and cadmium on alpha-synuclein aggregation in vitro and in cells. Int J Mol Sci 22:11455
Takaine M, Imamura H, Yoshida S (2022) High and stable ATP levels prevent aberrant intracellular protein aggregation in yeast. Elife 11:e67659
Vermulst M, Denney AS, Lang MJ, Hung CW, Moore S, Moseley MA, Thompson JW, Madden V, Gauer J, Wolfe KJ, Summers DW, Schleit J, Sutphin GL, Haroon S, Holczbauer A, Caine J, Jorgenson J, Cyr D, Kaeberlein M, Strathern JN, Duncan MC, Erie DA (2015) Transcription errors induce proteotoxic stress and shorten cellular lifespan. Nat Commun 6:8065
Francisco S, Ferreira M, Moura G, Soares AR, Santos MAS (2020) Does proteostasis get lost in translation? Implications for protein aggregation across the lifespan. Ageing Res Rev 62:101119
Lau A, Zheng Y, Tao S, Wang H, Whitman SA, White E, Zhang DD (2013) Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol Cell Biol 33:2436–2446
Dodson M, Liu P, Jiang T, Ambrose AJ, Luo G, Rojo de la Vega M, Cholanians AB, Wong PK, Chapman E, Zhang DD (2018) Increased O-GlcNAcylation of SNAP29 drives arsenic-induced autophagic dysfunction. Mol Cell Biol 38:e00595-e617
Tam LM, Wang Y (2020) Arsenic exposure and compromised protein quality control. Chem Res Toxicol 33:1594–1604
Tillotson J, Zerio CJ, Harder B, Ambrose AJ, Jung KS, Kang M, Zhang DD, Chapman E (2017) Arsenic compromises Both p97 and proteasome functions. Chem Res Toxicol 30:1508–1514
Kirkpatrick DS, Dale KV, Catania JM, Gandolfi AJ (2003) Low-level arsenite causes accumulation of ubiquitinated proteins in rabbit renal cortical slices and HEK293 cells. Toxicol Appl Pharmacol 186:101–109
Lee D, Goldberg AL (2022) 26S proteasomes become stably activated upon heat shock when ubiquitination and protein degradation increase. Proc Natl Acad Sci U S A 119:e2122482119
Jacobson T, Priya S, Sharma SK, Andersson S, Jakobsson S, Tanghe R, Ashouri A, Rauch S, Goloubinoff P, Christen P, Tamás MJ (2017) Cadmium causes misfolding and aggregation of cytosolic proteins in yeast. Mol Cell Biol 37:e00490-e516
Tamás MJ, Fauvet B, Christen P, Goloubinoff P (2018) Misfolding and aggregation of nascent proteins: a novel mode of toxic cadmium action in vivo. Curr Genet 64:177–181
Zuily L, Lahrach N, Fassler R, Genest O, Faller P, Seneque O, Denis Y, Castanie-Cornet MP, Genevaux P, Jakob U, Reichmann D, Giudici-Orticoni MT, Ilbert M (2022) Copper induces protein aggregation, a toxic process compensated by molecular chaperones. MBio 13:e0325121
Holland S, Lodwig E, Sideri T, Reader T, Clarke I, Gkargkas K, Hoyle DC, Delneri D, Oliver SG, Avery SV (2007) Application of the comprehensive set of heterozygous yeast deletion mutants to elucidate the molecular basis of cellular chromium toxicity. Genome Biol 8:R268
Sharma SK, Goloubinoff P, Christen P (2008) Heavy metal ions are potent inhibitors of protein folding. Biochem Biophys Res Commun 372:341–345
Brochu C, Haimeur A, Ouellette M (2004) The heat shock protein HSP70 and heat shock cognate protein HSC70 contribute to antimony tolerance in the protozoan parasite leishmania. Cell Stress Chaperones 9:294–303
Verdugo M, Ruiz Encinar J, Costa-Fernandez JM, Menendez-Miranda M, Bouzas-Ramos D, Bravo M, Quiroz W (2017) Study of conformational changes and protein aggregation of bovine serum albumin in presence of Sb(III) and Sb(V). PLoS ONE 12:e0170869
Gu JH, Lin DD, Sun YY, Guo YZ, Chen B, Zhang YM, Liu FS (2022) Integrating transcriptome and physiological analysis to reveal the essential responses of Daphnia magna to antimony trioxide nanoparticle. J Hazard Mater 437:129303
Alberti S, Hyman AA (2021) Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol 22:196–213
Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18:285–298
Wang B, Zhang L, Dai T, Qin Z, Lu H, Zhang L, Zhou F (2021) Liquid-liquid phase separation in human health and diseases. Signal Transduct Target Ther 6:290
Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P (2005) Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169:871–884
Kedersha NL, Gupta M, Li W, Miller I, Anderson P (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147:1431–1442
Protter DSW, Parker R (2016) Principles and properties of stress granules. Trends Cell Biol 26:668–679
Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, Ivanov P (2017) Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci 130:927–937
Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B (2013) Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol 23:2452–2462
Lallemand-Breitenbach V, de The H (2010) PML nuclear bodies. CSH Perspect Biol 2:a000661
Zhu J, Koken MH, Quignon F, Chelbi-Alix MK, Degos L, Wang ZY, Chen Z, de The H (1997) Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc Natl Acad Sci U S A 94:3978–3983
Mediani L, Guillen-Boixet J, Vinet J, Franzmann TM, Bigi I, Mateju D, Carra AD, Morelli FF, Tiago T, Poser I, Alberti S, Carra S (2019) Defective ribosomal products challenge nuclear function by impairing nuclear condensate dynamics and immobilizing ubiquitin. EMBO J 38:e101341
Spegg V, Altmeyer M (2021) Biomolecular condensates at sites of DNA damage: more than just a phase. DNA Repair 106:103179
Oshidari R, Huang R, Medghalchi M, Tse EYW, Ashgriz N, Lee HO, Wyatt H, Mekhail K (2020) DNA repair by Rad52 liquid droplets. Nat Commun 11:695
Mine-Hattab J, Heltberg M, Villemeur M, Guedj C, Mora T, Walczak AM, Dahan M, Taddei A (2021) Single molecule microscopy reveals key physical features of repair foci in living cells. Elife 10:e60577
Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M (2019) Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J 38:e101379
Pessina F, Giavazzi F, Yin YD, Gioia U, Vitelli V, Galbiati A, Barozzi S, Garre M, Oldani A, Flaus A, Cerbino R, Parazzoli D, Rothenberg E, di Fagagna FD (2019) Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol 21:1286–1299
Spannl S, Tereshchenko M, Mastromarco GJ, Ihn SJ, Lee HO (2019) Biomolecular condensates in neurodegeneration and cancer. Traffic 20:890–911
Volkov VA, Akhmanova A (2023) Phase separation on microtubules: from droplet formation to cellular function? Trends Cell Biol. https://doi.org/10.1016/j.tcb.2023.06.004
Povarova OI, Antifeeva IA, Fonin AV, Turoverov KK, Kuznetsova IM (2023) The role of liquid–liquid phase separation in actin polymerization. Int J Mol Sci 24:3281
Zhang K, Huang MD, Li A, Wen J, Yan LL, Li YH, Guo LM, Senthil KS, Zhou YY, Chen GB, Liu Y, Zhang XF, Yao XL, Qin DJ, Su HX (2023) DIAPH3 condensates formed by liquid–liquid phase separation act as a regulatory hub for stress-induced actin cytoskeleton remodeling. Cell Rep 42:111986
Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grofte M, Rask MBD, Streicher W, Jungmichel S, Nielsen ML, Lukas J (2015) Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun 6:8088
Jack A, Kim Y, Strom AR, Lee DSW, Williams B, Schaub JM, Kellogg EH, Finkelstein IJ, Ferro LS, Yildiz A, Brangwynne CP (2022) Compartmentalization of telomeres through DNA-scaffolded phase separation. Dev Cell 57:277-290.e9
Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, Abraham BJ, Afeyan LK, Guo YE, Rimel JK, Fant CB, Schuijers J, Lee TI, Taatjes DJ, Young RA (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175:1842-1855.e16
Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, Cisse II (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361:412–415
Chowdhary S, Kainth AS, Paracha S, Gross DS, Pincus D (2022) Inducible transcriptional condensates drive 3D genome reorganization in the heat shock response. Mol Cell 82:4386-4399.e7
Jochem M, Ende L, Isasa M, Ang J, Schnell H, Guerra-Moreno A, Micoogullari Y, Bhanu M, Gygi SP, Hanna J (2019) Targeted degradation of glucose transporters protects against arsenic toxicity. Mol Cell Biol 39:e00559-e618
Wysocki R, Chéry CC, Wawrzycka D, Van Hulle M, Cornelis R, Thevelein JM, Tamás MJ (2001) The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Mol Microbiol 40:1391–1401
Talemi SR, Jacobson T, Garla V, Navarrete C, Wagner A, Tamás MJ, Schaber J (2014) Mathematical modelling of arsenic transport, distribution and detoxification processes in yeast. Mol Microbiol 92:1343–1356
Thorsen M, Lagniel G, Kristiansson E, Junot C, Nerman O, Labarre J, Tamás MJ (2007) Quantitative transcriptome, proteome, and sulfur metabolite profiling of the Saccharomyces cerevisiae response to arsenite. Physiol Genomics 30:35–43
Liu B, Han Y, Qian SB (2013) Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol Cell 49:453–463
Brostrom CO, Prostko CR, Kaufman RJ, Brostrom MA (1996) Inhibition of translational initiation by activators of the glucose-regulated stress protein and heat shock protein stress response systems. Role of the interferon-inducible double-stranded RNA-activated eukaryotic initiation factor 2alpha kinase. J Biol Chem 271:24995–25002
Haugen AC, Kelley R, Collins JB, Tucker CJ, Deng C, Afshari CA, Brown JM, Ideker T, Van Houten B (2004) Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol 5:R95
Hosiner D, Lempiainen H, Reiter W, Urban J, Loewith R, Ammerer G, Schweyen R, Shore D, Schuller C (2009) Arsenic toxicity to Saccharomyces cerevisiae is a consequence of inhibition of the TORC1 kinase combined with a chronic stress response. Mol Biol Cell 20:1048–1057
Johnston D, Oppermann H, Jackson J, Levinson W (1980) Induction of four proteins in chick embryo cells by sodium arsenite. J Biol Chem 255:6975–6980
Levinson W, Oppermann H, Jackson J (1980) Transition series metals and sulfhydryl reagents induce the synthesis of four proteins in eukaryotic cells. Biochim Biophys Acta 606:170–180
Zheng PZ, Wang KK, Zhang QY, Huang QH, Du YZ, Zhang QH, Xiao DK, Shen SH, Imbeaud S, Eveno E, Zhao CJ, Chen YL, Fan HY, Waxman S, Auffray C, Jin G, Chen SJ, Chen Z, Zhang J (2005) Systems analysis of transcriptome and proteome in retinoic acid/arsenic trioxide-induced cell differentiation/apoptosis of promyelocytic leukemia. Proc Natl Acad Sci U S A 102:7653–7658
Yun C, Stanhill A, Yang Y, Zhang Y, Haynes CM, Xu CF, Neubert TA, Mor A, Philips MR, Ron D (2008) Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc Natl Acad Sci U S A 105:7094–7099
Turakhiya A, Meyer SR, Marincola G, Bohm S, Vanselow JT, Schlosser A, Hofmann K, Buchberger A (2018) ZFAND1 recruits p97 and the 26S proteasome to promote the clearance of arsenite-induced stress granules. Mol Cell 70:906-919.e7
Guerra-Moreno A, Hanna J (2016) Tmc1 is a dynamically regulated effector of the Rpn4 proteotoxic stress response. J Biol Chem 291:14788–14795
Yau R, Rape M (2016) The increasing complexity of the ubiquitin code. Nat Cell Biol 18:579–586
Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81:203–229
Meyer HJ, Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157:910–921
Ohtake F, Tsuchiya H, Saeki Y, Tanaka K (2018) K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc Natl Acad Sci U S A 115:E1401–E1408
Zhang T, Qi Y, Liao M, Xu M, Bower KA, Frank JA, Shen HM, Luo J, Shi X, Chen G (2012) Autophagy is a cell self-protective mechanism against arsenic-induced cell transformation. Toxicol Sci 130:298–308
Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, Airoldi EM, Pan T, Budnik BA, Drummond DA (2015) Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162:1286–1298
Sanchez Y, Lindquist SL (1990) HSP104 required for induced thermotolerance. Science 248:1112–1115
Sanchez Y, Taulien J, Borkovich KA, Lindquist S (1992) Hsp104 is required for tolerance to many forms of stress. EMBO J 11:2357–2364
Chamera T, Klosowska A, Janta A, Wyszkowski H, Obuchowski I, Gumowski K, Liberek K (2019) Selective Hsp70-dependent docking of Hsp104 to protein aggregates protects the cell from the toxicity of the disaggregase. J Mol Biol 431:2180–2196
Lee DH, Sherman MY, Goldberg AL (1996) Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol 16:4773–4781
Lee DH, Sherman MY, Goldberg AL (2016) The requirements of yeast Hsp70 of SSA family for the ubiquitin-dependent degradation of short-lived and abnormal proteins. Biochem Biophys Res Commun 475:100–106
Fang NN, Chan GT, Zhu M, Comyn SA, Persaud A, Deshaies RJ, Rotin D, Gsponer J, Mayor T (2014) Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress. Nat Cell Biol 16:1227–1237
Shiber A, Breuer W, Brandeis M, Ravid T (2013) Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol Biol Cell 24:2076–2087
Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU (2013) Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82:323–355
Comyn SA, Young BP, Loewen CJ, Mayor T (2016) Prefoldin promotes proteasomal degradation of cytosolic proteins with missense mutations by maintaining substrate solubility. PLoS Genet 12:e1006184
Kaganovich D, Kopito R, Frydman J (2008) Misfolded proteins partition between two distinct quality control compartments. Nature 454:1088–1095
Specht S, Miller SB, Mogk A, Bukau B (2011) Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol 195:617–629
Escusa-Toret S, Vonk WI, Frydman J (2013) Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol 15:1231–1243
Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S (2012) Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell 23:3041–3056
Soto C, Pritzkow S (2018) Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat Neurosci 21:1332–1340
Wilson DM 3rd, Cookson MR, Van Den Bosch L, Zetterberg H, Holtzman DM, Dewachter I (2023) Hallmarks of neurodegenerative diseases. Cell 186:693–714
Dani SU (2010) Arsenic for the fool: an exponential connection. Sci Total Environ 408:1842–1846
Elbaz A, Clavel J, Rathouz PJ, Moisan F, Galanaud JP, Delemotte B, Alperovitch A, Tzourio C (2009) Professional exposure to pesticides and Parkinson disease. Ann Neurol 66:494–504
Gong G, O’Bryant SE (2010) The arsenic exposure hypothesis for Alzheimer disease. Alzheimer Dis Assoc Disord 24:311–316
Li XL, Zhan RQ, Zheng W, Jiang H, Zhang DF, Shen XL (2020) Positive association between soil arsenic concentration and mortality from Alzheimer’s disease in mainland China. J Trace Elem Med Biol 59:126452
Lee CP, Zhu CH, Su CC (2021) Increased prevalence of Parkinson’s disease in soils with high arsenic levels. Parkinsonism Relat Disord 88:19–23
Tyler CR, Allan AM (2014) The effects of arsenic exposure on neurological and cognitive dysfunction in human and rodent studies: a review. Curr Environ Health Rep 1:132–147
Sharma A, Kumar S (2019) Arsenic exposure with reference to neurological impairment: an overview. Rev Environ Health 34:403–414
Magalhaes P, Lashuel HA (2022) Opportunities and challenges of alpha-synuclein as a potential biomarker for Parkinson’s disease and other synucleinopathies. NPJ Parkinsons Dis 8:93
Lin AM, Chao PL, Fang SF, Chi CW, Yang CH (2007) Endoplasmic reticulum stress is involved in arsenite-induced oxidative injury in rat brain. Toxicol Appl Pharmacol 224:138–146
Cholanians AB, Phan AV, Ditzel EJ, Camenisch TD, Lau SS, Monks TJ (2016) From the cover: arsenic induces accumulation of alpha-synuclein: implications for synucleinopathies and neurodegeneration. Toxicol Sci 153:271–281
Drobna Z (2020) Activation of Lrrk2 and alpha-synuclein in substantia nigra, striatum, and cerebellum after chronic exposure to arsenite. Toxicol Appl Pharmacol 408:115278
Marchand A, Drouyer M, Sarchione A, Chartier-Harlin MC, Taymans JM (2020) LRRK2 phosphorylation, more than an epiphenomenon. Front Neurosci 14:527
Oueslati A, Fournier M, Lashuel HA (2010) Role of post-translational modifications in modulating the structure, function and toxicity of alpha-synuclein: implications for Parkinson’s disease pathogenesis and therapies. Prog Brain Res 183:115–145
Byrd EJ, Wilkinson M, Radford SE, Sobott F (2023) Taking charge: metal ions accelerate amyloid aggregation in sequence variants of alpha-synuclein. J Am Soc Mass Spectrom 34:493–504
Binolfi A, Rasia RM, Bertoncini CW, Ceolin M, Zweckstetter M, Griesinger C, Jovin TM, Fernandez CO (2006) Interaction of alpha-synuclein with divalent metal ions reveals key differences: a link between structure, binding specificity and fibrillation enhancement. J Am Chem Soc 128:9893–9901
Carboni E, Lingor P (2015) Insights on the interaction of alpha-synuclein and metals in the pathophysiology of Parkinson’s disease. Metallomics 7:395–404
Uversky VN, Li J, Fink AL (2001) Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson’s disease and heavy metal exposure. J Biol Chem 276:44284–44296
Silva MVF, Loures CMG, Alves LCV, de Souza LC, Borges KBG, Carvalho MDG (2019) Alzheimer’s disease: risk factors and potentially protective measures. J Biomed Sci 26:33
Busche MA, Hyman BT (2020) Synergy between amyloid-beta and tau in Alzheimer’s disease. Nat Neurosci 23:1183–1193
Zarazua S, Burger S, Delgado JM, Jimenez-Capdeville ME, Schliebs R (2011) Arsenic affects expression and processing of amyloid precursor protein (APP) in primary neuronal cells overexpressing the Swedish mutation of human APP. Int J Dev Neurosci 29:389–396
Ashok A, Rai NK, Tripathi S, Bandyopadhyay S (2015) Exposure to As-, Cd-, and Pb-mixture induces Abeta, amyloidogenic APP processing and cognitive impairments via oxidative stress-dependent neuroinflammation in young rats. Toxicol Sci 143:64–80
Nino SA, Morales-Martinez A, Chi-Ahumada E, Carrizales L, Salgado-Delgado R, Perez-Severiano F, Diaz-Cintra S, Jimenez-Capdeville ME, Zarazua S (2019) Arsenic exposure contributes to the bioenergetic damage in an Alzheimer’s disease model. ACS Chem Neurosci 10:323–336
Wisessaowapak C, Visitnonthachai D, Watcharasit P, Satayavivad J (2021) Prolonged arsenic exposure increases tau phosphorylation in differentiated SH-SY5Y cells: the contribution of GSK3 and ERK1/2. Environ Toxicol Pharmacol 84:103626
Giasson BI, Sampathu DM, Wilson CA, Vogelsberg-Ragaglia V, Mushynski WE, Lee VM (2002) The environmental toxin arsenite induces tau hyperphosphorylation. Biochemistry 41:15376–15387
Wisessaowapak C, Worasuttayangkurn L, Maliphol K, Nakareangrit W, Cholpraipimolrat W, Nookabkaew S, Watcharasit P, Satayavivad J (2022) The 28-day repeated arsenic exposure increases tau phosphorylation in the rat brain. Environ Toxicol Pharmacol 95:103974
Alonso A, Zaidi T, Novak M, Grundke-Iqbal I, Iqbal K (2001) Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proc Natl Acad Sci U S A 98:6923–6928
Iqbal K, Gong CX, Liu F (2013) Hyperphosphorylation-induced tau oligomers. Front Neurol 4:112
Tanu T, Anjum A, Jahan M, Nikkon F, Hoque M, Roy AK, Haque A, Himeno S, Hossain K, Saud ZA (2018) Antimony-induced neurobehavioral and biochemical perturbations in mice. Biol Trace Elem Res 186:199–207
Wang X, Zhu P, Xu S, Liu Y, Jin Y, Yu S, Wei H, Li J, Zhang Q, Hasegawa T, Yao C, Yoshimura H, Wu Q, Zhao X (2019) Antimony, a novel nerve poison, triggers neuronal autophagic death via reactive oxygen species-mediated inhibition of the protein kinase B/mammalian target of rapamycin pathway. Int J Biochem Cell Biol 114:105561
Xu S, Yang Z, Zhi Y, Yu S, Zhang T, Jiang J, Tang J, He H, Lu M, Wang X, Wu Q, Zhao X (2021) The effects of antimony on Alzheimer’s disease-like pathological changes in mice brain. Sci Total Environ 760:143235
Ainslie A, Huiting W, Barazzuol L, Bergink S (2021) Genome instability and loss of protein homeostasis: converging paths to neurodegeneration? Open Biol 11:200296
Negrini S, Gorgoulis VG, Halazonetis TD (2010) Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11:220–228
Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G (2023) Hallmarks of aging: an expanding universe. Cell 186:243–278
Yang JL, Weissman L, Bohr VA, Mattson MP (2008) Mitochondrial DNA damage and repair in neurodegenerative disorders. DNA Repair (Amst) 7:1110–1120
Coppede F, Migliore L (2015) DNA damage in neurodegenerative diseases. Mutat Res 776:84–97
Maiuri T, Suart CE, Hung CLK, Graham KJ, Barba Bazan CA, Truant R (2019) DNA Damage repair in Huntington’s disease and other neurodegenerative diseases. Neurotherapeutics 16:948–956
Hinkle JT, Patel J, Panicker N, Karuppagounder SS, Biswas D, Belingon B, Chen R, Brahmachari S, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM (2022) STING mediates neurodegeneration and neuroinflammation in nigrostriatal alpha-synucleinopathy. Proc Natl Acad Sci U S A 119:e2118819119
Suberbielle E, Djukic B, Evans M, Kim DH, Taneja P, Wang X, Finucane M, Knox J, Ho K, Devidze N, Masliah E, Mucke L (2015) DNA repair factor BRCA1 depletion occurs in Alzheimer brains and impairs cognitive function in mice. Nat Commun 6:8897
Sepe S, Milanese C, Gabriels S, Derks KW, Payan-Gomez C, van Wildfred IF, Rijksen YM, Nigg AL, Moreno S, Cerri S, Blandini F, Hoeijmakers JH, Mastroberardino PG (2016) Inefficient DNA repair is an aging-related modifier of Parkinson’s disease. Cell Rep 15:1866–1875
Mitra J, Guerrero EN, Hegde PM, Liachko NF, Wang H, Vasquez V, Gao J, Pandey A, Taylor JP, Kraemer BC, Wu P, Boldogh I, Garruto RM, Mitra S, Rao KS, Hegde ML (2019) Motor neuron disease-associated loss of nuclear TDP-43 is linked to DNA double-strand break repair defects. Proc Natl Acad Sci U S A 116:4696–4705
SenGupta T, Palikaras K, Esbensen YQ, Konstantinidis G, Galindo FJN, Achanta K, Kassahun H, Stavgiannoudaki I, Bohr VA, Akbari M, Gaare J, Tzoulis C, Tavernarakis N, Nilsen H (2021) Base excision repair causes age-dependent accumulation of single-stranded DNA breaks that contribute to Parkinson disease pathology. Cell Rep 36:109668
Thadathil N, Delotterie DF, Xiao J, Hori R, McDonald MP, Khan MM (2021) DNA double-strand break accumulation in Alzheimer’s disease: evidence from experimental models and postmortem human brains. Mol Neurobiol 58:118–131
Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204
Welch G, Tsai LH (2022) Mechanisms of DNA damage-mediated neurotoxicity in neurodegenerative disease. EMBO Rep 23:e54217
Lee JH, Mand MR, Kao CH, Zhou Y, Ryu SW, Richards AL, Coon JJ, Paull TT (2018) ATM directs DNA damage responses and proteostasis via genetically separable pathways. Sci Signal. 11:eaan5598
Lee JH, Ryu SW, Ender NA, Paull TT (2021) Poly-ADP-ribosylation drives loss of protein homeostasis in ATM and Mre11 deficiency. Mol Cell 81:1515-1533.e5
Huiting W, Dekker SL, van der Lienden JCJ, Mergener R, Musskopf MK, Furtado GV, Gerrits E, Coit D, Oghbaie M, Di Stefano LH, Schepers H, van Waarde-Verhagen M, Couzijn S, Barazzuol L, LaCava J, Kampinga HH, Bergink S (2022) Targeting DNA topoisomerases or checkpoint kinases results in an overload of chaperone systems, triggering aggregation of a metastable subproteome. Elife 11:e70726
Corcoles-Saez I, Dong K, Johnson AL, Waskiewicz E, Costanzo M, Boone C, Cha RS (2018) Essential function of Mec1, the budding yeast ATM/ATR checkpoint-response kinase protein homeostasis. Dev Cell 46(495–503):e2
Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133
Wang H, Guo W, Mitra J, Hegde PM, Vandoorne T, Eckelmann BJ, Mitra S, Tomkinson AE, Van Den Bosch L, Hegde ML (2018) Mutant FUS causes DNA ligation defects to inhibit oxidative damage repair in amyotrophic lateral sclerosis. Nat Commun 9:3683
Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH (2013) Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci 16:1383–1391
Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS (2013) The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem 288:24731–24741
Maiuri T, Mocle AJ, Hung CL, Xia J, van Roon-Mom WM, Truant R (2017) Huntingtin is a scaffolding protein in the ATM oxidative DNA damage response complex. Hum Mol Genet 26:395–406
Gao R, Chakraborty A, Geater C, Pradhan S, Gordon KL, Snowden J, Yuan S, Dickey AS, Choudhary S, Ashizawa T, Ellerby LM, La Spada AR, Thompson LM, Hazra TK, Sarkar PS (2019) Mutant huntingtin impairs PNKP and ATXN3, disrupting DNA repair and transcription. Elife 8:e42988
Schaser AJ, Osterberg VR, Dent SE, Stackhouse TL, Wakeham CM, Boutros SW, Weston LJ, Owen N, Weissman TA, Luna E, Raber J, Luk KC, McCullough AK, Woltjer RL, Unni VK (2019) Alpha-synuclein is a DNA binding protein that modulates DNA repair with implications for Lewy body disorders. Sci Rep 9:10919
Chaudhary AR, Berger F, Berger CL, Hendricks AG (2018) Tau directs intracellular trafficking by regulating the forces exerted by kinesin and dynein teams. Traffic 19:111–121
Rico T, Denechaud M, Caillierez R, Comptdaer T, Adriaenssens E, Buee L, Lefebvre B (2022) Cancer cells upregulate tau to gain resistance to DNA damaging agents. Cancers (Basel). 15:116
Colnaghi L, Rondelli D, Muzi-Falconi M, Sertic S (2020) Tau and DNA damage in neurodegeneration. Brain Sci 10:946
Vasquez V, Mitra J, Hegde PM, Pandey A, Sengupta S, Mitra S, Rao KS, Hegde ML (2017) Chromatin-bound oxidized alpha-synuclein causes strand breaks in neuronal genomes in in vitro models of Parkinson’s disease. J Alzheimers Dis 60:S133–S150
Kam TI, Mao X, Park H, Chou SC, Karuppagounder SS, Umanah GE, Yun SP, Brahmachari S, Panicker N, Chen R, Andrabi SA, Qi C, Poirier GG, Pletnikova O, Troncoso JC, Bekris LM, Leverenz JB, Pantelyat A, Ko HS, Rosenthal LS, Dawson TM, Dawson VL (2018) Poly(ADP-ribose) drives pathologic alpha-synuclein neurodegeneration in Parkinson’s disease. Science 362:eaat8407
Duan Y, Du A, Gu J, Duan G, Wang C, Gui X, Ma Z, Qian B, Deng X, Zhang K, Sun L, Tian K, Zhang Y, Jiang H, Liu C, Fang Y (2019) PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res 29:233–247
Cardinale A, Paldino E, Giampa C, Bernardi G, Fusco FR (2015) PARP-1 inhibition is neuroprotective in the R6/2 mouse model of Huntington’s disease. PLoS ONE 10:e0134482
Nakamura M, Kaneko S, Dickson DW, Kusaka H (2020) Aberrant accumulation of BRCA1 in Alzheimer disease and other tauopathies. J Neuropathol Exp Neurol 79:22–33
Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA (2004) Gene regulation and DNA damage in the ageing human brain. Nature 429:883–891
Lin MT, Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443:787–795
Liu Z, Zhou T, Ziegler AC, Dimitrion P, Zuo L (2017) Oxidative stress in neurodegenerative diseases: from molecular mechanisms to clinical applications. Ox Med Cell Longev 2017:2525967
Nakabeppu Y, Tsuchimoto D, Yamaguchi H, Sakumi K (2007) Oxidative damage in nucleic acids and Parkinson’s disease. J Neurosci Res 85:919–934
Wang ZX, Li YL, Pu JL, Zhang BR (2023) DNA damage-mediated neurotoxicity in Parkinson’s disease. Int J Mol Sci 24:6313
Pollari E, Goldsteins G, Bart G, Koistinaho J, Giniatullin R (2014) The role of oxidative stress in degeneration of the neuromuscular junction in amyotrophic lateral sclerosis. Front Cell Neurosci 8:131
Funding
Open access funding provided by University of Gothenburg. This work was supported by grants from the National Science Centre, Poland (grant number 2016/21/B/NZ2/01751) to R.W. and the Swedish Research Council (grant number 2018–03577) to M.J.T.
Author information
Authors and Affiliations
Contributions
RW and MJT contributed to the study conception and design. JIR prepared all figures. All authors contributed to literature search, writing of the first draft of the manuscript, and commenting on previous versions of the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wysocki, R., Rodrigues, J.I., Litwin, I. et al. Mechanisms of genotoxicity and proteotoxicity induced by the metalloids arsenic and antimony. Cell. Mol. Life Sci. 80, 342 (2023). https://doi.org/10.1007/s00018-023-04992-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04992-5