Abstract
Objective
Intriguingly, hyperinsulinemia, and hyperglycemia can predispose insulin resistance, obesity, and type 2 diabetes, leading to metabolic disturbances. Conversely, physical exercise stimulates skeletal muscle glucose uptake, improving whole-body glucose homeostasis. Therefore, we investigated the impact of short-term physical activity in a mouse model (Slc2a4+/−) that spontaneously develops hyperinsulinemia and hyperglycemia even when fed on a chow diet.
Methods
Slc2a4+/− mice were used, that performed 5 days of endurance or strength exercise training. Further analysis included physiological tests (GTT and ITT), skeletal muscle glucose uptake, skeletal muscle RNA-sequencing, mitochondrial function, and experiments with C2C12 cell line.
Results
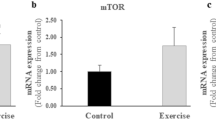
When Slc2a4+/− mice were submitted to the endurance or strength training protocol, improvements were observed in the skeletal muscle glucose uptake and glucose metabolism, associated with broad transcriptomic modulation, that was, in part, related to mitochondrial adaptations. The endurance training, but not the strength protocol, was effective in improving skeletal muscle mitochondrial activity and unfolded protein response markers (UPRmt). Moreover, experiments with C2C12 cells indicated that insulin or glucose levels could contribute to these mitochondrial adaptations in skeletal muscle.
Conclusions
Both short-term exercise protocols were efficient in whole-body glucose homeostasis and insulin resistance. While endurance exercise plays an important role in transcriptome and mitochondrial activity, strength exercise mostly affects post-translational mechanisms and protein synthesis in skeletal muscle. Thus, the performance of both types of physical exercise proved to be a very effective way to mitigate the impacts of hyperglycemia and hyperinsulinemia in the Slc2a4+/− mouse model.






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Wiebe N, Ye F, Crumley ET, Bello A, Stenvinkel P, Tonelli M (2021) Temporal associations among body mass index, fasting insulin, and systemic inflammation. JAMA Netw Open 4(3):e211263. https://doi.org/10.1001/jamanetworkopen.2021.1263
Dankner R, Chetrit A, Shanik MHH, Raz I, Roth J (2012) Basal state hyperinsulinemia in healthy normoglycemic adults heralds dysglycemia after more than two decades of follow up. Diabetes Metab Res Rev 28(7):618–624. https://doi.org/10.1002/dmrr.2322
Zimmet PZ, Collins VR, Dowse GK, Knight LT (1992) Hyperinsulinaemia in youth is a predictor of type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia 35(6):534–541. https://doi.org/10.1007/BF00400481
Rothman DL, Magnusson I, Cline G, Gerard D, Kahn CR, Shulman RG et al (1995) Decreased muscle glucose transport/phosphorylation is an early defect in the pathogenesis of non-insulin-dependent diabetes mellitus. Proc Natl Acad Sci 92(4):983–987. https://doi.org/10.1073/pnas.92.4.983
DeFronzo RA, Gunnarsson R, Björkman O, Olsson M, Wahren J (1985) Effects of insulin on peripheral and splanchnic glucose metabolism in noninsulin-dependent (type II) diabetes mellitus. J Clin Investig 76(1):149–155. https://doi.org/10.1172/JCI111938
DeFronzo RA, Jacot E, Jequier E, Maeder E, Wahren J, Felber JP (1981) The effect of insulin on the disposal of intravenous glucose: results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes 30(12):1000–1007. https://doi.org/10.2337/diab.30.12.1000
Marangou AG, Weber KM, Boston RC, Aitken PM, Heggie JCP, Kirsner RLG et al (1986) Metabolic consequences of prolonged hyperinsulinemia in humans: evidence for induction of insulin insensitivity. Diabetes 35(12):1383–1389. https://doi.org/10.2337/diab.35.12.1383
Cen HH, Hussein B, Botezelli JD, Wang S, Zhang JA, Noursadeghi N et al (2022) Human and mouse muscle transcriptomic analyses identify insulin receptor mRNA downregulation in hyperinsulinemia-associated insulin resistance. FASEB J. https://doi.org/10.1096/fj.202100497RR
Page MM, Skovs S, Cen H, Chiu AP, Dionne DA, Hutchinson DF et al (2018) Reducing insulin via conditional partial gene ablation in adults reverses diet-induced weight gain. FASEB J 32(3):1196–1206. https://doi.org/10.1096/fj.201700518R
Templeman NM, Flibotte S, Chik JHL, Sinha S, Lim GE, Foster LJ et al (2017) Reduced circulating insulin enhances insulin sensitivity in old mice and extends lifespan. Cell Rep 20(2):451–463. https://doi.org/10.1016/j.celrep.2017.06.048
Mehran AE, Templeman NM, Brigidi GS, Lim GE, Chu K-Y, Hu X et al (2012) Hyperinsulinemia drives diet-induced obesity independently of brain insulin production. Cell Metab 16(6):723–737. https://doi.org/10.1016/j.cmet.2012.10.019
Huang S, Czech MP (2007) The GLUT4 glucose transporter. Cell Metab 5(4):237–252. https://doi.org/10.1016/j.cmet.2007.03.006
Reno CM, Puente EC, Sheng Z, Daphna-Iken D, Bree AJ, Routh VH et al (2017) Brain GLUT4 knockout mice have impaired glucose tolerance, decreased insulin sensitivity, and impaired hypoglycemic counterregulation. Diabetes 66(3):587–597. https://doi.org/10.2337/db16-0917
Sylow L, Kleinert M, Richter EA, Jensen TE (2017) Exercise-stimulated glucose uptake—regulation and implications for glycaemic control. Nat Rev Endocrinol 13(3):133–148. https://doi.org/10.1038/nrendo.2016.162
Sangwung P, Petersen KF, Shulman GI, Knowles JW (2020) Mitochondrial dysfunction, insulin resistance, and potential genetic implications. Endocrinology. https://doi.org/10.1210/endocr/bqaa017
Mogensen M, Sahlin K, Fernström M, Glintborg D, Vind BF, Beck-Nielsen H et al (2007) Mitochondrial respiration is decreased in skeletal muscle of patients with type 2 diabetes. Diabetes 56(6):1592–1599. https://doi.org/10.2337/db06-0981
Stenbit AE, Tsao TS, Li J, Burcelin R, Geenen DL, Factor SM et al (1997) GLUT4 heterozygous knockout mice develop muscle insulin resistance and diabetes. Nat Med 3(10):1096–1101
Hoene M, Kappler L, Kollipara L, Hu C, Irmler M, Bleher D et al (2021) Exercise prevents fatty liver by modifying the compensatory response of mitochondrial metabolism to excess substrate availability. Mol Metab 54:101359. https://doi.org/10.1016/j.molmet.2021.101359
Fredrickson G, Barrow F, Dietsche K, Parthiban P, Khan S, Robert S et al (2021) Exercise of high intensity ameliorates hepatic inflammation and the progression of NASH. Mol Metab. https://doi.org/10.1016/j.molmet.2021.101270
Macfarlane DP, Forbes S, Walker BR (2008) Glucocorticoids and fatty acid metabolism in humans: fuelling fat redistribution in the metabolic syndrome. J Endocrinol 197(2):189–204. https://doi.org/10.1677/JOE-08-0054
Hawley JAA, Hargreaves M, Joyner MJJ, Zierath JRR (2014) Integrative biology of exercise. Cell 159(4):738–749. https://doi.org/10.1016/j.cell.2014.10.029
Komada M, Takao K, Miyakawa T (2008) Elevated Plus Maze for Mice. J Vis Exp. https://doi.org/10.3791/1088
Oliveira de Almeida WA, Maculano Esteves A, Leite de Almeida-Júnior C, Lee KS, Kannebley Frank M, Oliveira Mariano M et al (2014) The effects of long-term dopaminergic treatment on locomotor behavior in rats. Sleep Sci 7(4):203–208. https://doi.org/10.1016/j.slsci.2014.10.003
Bala M, Gupta P, Gupta S, Dua A, Injeti E, Mittal A (2021) Efficient and modified 2-NBDG assay to measure glucose uptake in cultured myotubes. J Pharmacol Toxicol Methods. https://doi.org/10.1016/j.vascn.2021.107069
Roden M, Shulman GI (2019) The integrative biology of type 2 diabetes. Nature 576(7785):51–60. https://doi.org/10.1038/s41586-019-1797-8
Shanik MH, Xu Y, Skrha J, Dankner R, Zick Y, Roth J (2008) Insulin resistance and hyperinsulinemia: is hyperinsulinemia the cart or the horse? Diabetes Care. https://doi.org/10.2337/dc08-s264
Corkey BE (2012) Banting lecture 2011: hyperinsulinemia: cause or consequence? Diabetes 61(1):4–13. https://doi.org/10.2337/db11-1483
Bird SR, Hawley JA (2017) Update on the effects of physical activity on insulin sensitivity in humans. BMJ Open Sport Exerc Med 2(1):e000143. https://doi.org/10.1136/bmjsem-2016-000143
Richter EA, Sylow L, Hargreaves M (2021) Interactions between insulin and exercise. Biochem J 478(21):3827–3846. https://doi.org/10.1042/BCJ20210185
Hu G, Rico-Sanz J, Lakka TA, Tuomilehto J (2006) Exercise, genetics and prevention of type 2 diabetes. Essays Biochem 42:177–192. https://doi.org/10.1042/bse0420177
Rome S, Clément K, Rabasa-Lhoret R, Loizon E, Poitou C, Barsh GS et al (2003) Microarray profiling of human skeletal muscle reveals that insulin regulates ∼800 genes during a hyperinsulinemic clamp. J Biol Chem 278(20):18063–18068. https://doi.org/10.1074/jbc.M300293200
Batista TM, Garcia-Martin R, Cai W, Konishi M, O’Neill BT, Sakaguchi M et al (2019) Multi-dimensional transcriptional remodeling by physiological insulin in vivo. Cell Rep 26(12):3429-3443. https://doi.org/10.1016/j.celrep.2019.02.081 (e3)
Wolfrum C, Besser D, Luca E, Stoffel M (2003) Insulin regulates the activity of forkhead transcription factor Hnf-3β/Foxa-2 by Akt-mediated phosphorylation and nuclear/cytosolic localization. Proc Natl Acad Sci 100(20):11624–11629. https://doi.org/10.1073/pnas.1931483100
Batista TM, Jayavelu AK, Wewer Albrechtsen NJ, Iovino S, Lebastchi J, Pan H et al (2020) A cell-autonomous signature of dysregulated protein phosphorylation underlies muscle insulin resistance in type 2 diabetes. Cell Metab 32(5):844-859. https://doi.org/10.1016/j.cmet.2020.08.007 (e5)
Sato S, Dyar KA, Treebak JT, Jepsen SL, Ehrlich AM, Ashcroft SP et al (2022) Atlas of exercise metabolism reveals time-dependent signatures of metabolic homeostasis. Cell Metab 34(2):329-345.e8. https://doi.org/10.1016/j.cmet.2021.12.016 (e8)
Srikanthan P, Karlamangla AS (2011) Relative muscle mass is inversely associated with insulin resistance and prediabetes. findings from the third national health and nutrition examination survey. J Clin Endocrinol Metab 96(9):2898–2903. https://doi.org/10.1210/jc.2011-0435
Kim K, Park SM (2018) Association of muscle mass and fat mass with insulin resistance and the prevalence of metabolic syndrome in Korean adults: a cross-sectional study. Sci Rep 8(1):2703. https://doi.org/10.1038/s41598-018-21168-5
MacDonald TL, Pattamaprapanont P, Cooney EM, Nava RC, Mitri J, Hafida S et al (2022) Canagliflozin prevents hyperglycemia-associated muscle extracellular matrix accumulation and improves the adaptive response to aerobic exercise. Diabetes 71(5):881–893. https://doi.org/10.2337/db21-0934
MacDonald TL, Pattamaprapanont P, Pathak P, Fernandez N, Freitas EC, Hafida S et al (2020) Hyperglycaemia is associated with impaired muscle signalling and aerobic adaptation to exercise. Nat Metab 2(9):902–917. https://doi.org/10.1038/s42255-020-0240-7
Anderson DM, Anderson KM, Chang C-L, Makarewich CA, Nelson BR, McAnally JR et al (2015) A micropeptide encoded by a putative long noncoding rna regulates muscle performance. Cell 160(4):595–606. https://doi.org/10.1016/j.cell.2015.01.009
Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB et al (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464(7285):121–125. https://doi.org/10.1038/nature08778
Jing E, O’Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S et al (2013) Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes 62(10):3404–3417. https://doi.org/10.2337/db12-1650
Koh J-H, Pataky MW, Dasari S, Klaus KA, Vuckovic I, Ruegsegger GN et al (2022) Enhancement of anaerobic glycolysis—a role of PGC-1α4 in resistance exercise. Nat Commun 13(1):2324. https://doi.org/10.1038/s41467-022-30056-6
Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G et al (2013) Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497(7450):451–457. https://doi.org/10.1038/nature12188
Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P et al (2016) NAD + repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352(6292):1436–1443. https://doi.org/10.1126/science.aaf2693
Pirinen E, Cantó C, Jo YS, Morato L, Zhang H, Menzies KJ et al (2014) Pharmacological inhibition of poly(ADP-Ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab 19(6):1034–1041. https://doi.org/10.1016/j.cmet.2014.04.002
van de Weijer T, Phielix E, Bilet L, Williams EG, Ropelle ER, Bierwagen A et al (2015) Evidence for a direct effect of the NAD+ precursor acipimox on muscle mitochondrial function in humans. Diabetes 64(4):1193–1201. https://doi.org/10.2337/db14-0667
Cordeiro AV, Peruca GF, Braga RR, Brícola RS, Lenhare L, Silva VRR et al (2021) High-intensity exercise training induces mitonuclear imbalance and activates the mitochondrial unfolded protein response in the skeletal muscle of aged mice. GeroScience 43(3):1513–1518. https://doi.org/10.1007/s11357-020-00246-5
Cordeiro AV, Brícola RS, Braga RR, Lenhare L, Silva VRR, Anaruma CP et al (2020) Aerobic Exercise Training Induces the Mitonuclear Imbalance and UPRmt in the Skeletal Muscle of Aged Mice. J Gerontol Series A 75(12):2258–2261. https://doi.org/10.1093/gerona/glaa059
Braga RR, Crisol BM, Brícola RS, Sant’ana, MR, Nakandakari SCBR, Costa SO et al (2021) Exercise alters the mitochondrial proteostasis and induces the mitonuclear imbalance and UPRmt in the hypothalamus of mice. Sci Rep. https://doi.org/10.1038/s41598-021-82352-8
Gao AW, El Alam G, Lalou A, Li TY, Molenaars M, Zhu Y et al (2022) Multi-omics analysis identifies essential regulators of mitochondrial stress response in two wild-type C. elegans strains. IScience 25(2):103734. https://doi.org/10.1016/j.isci.2022.103734
Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A et al (2015) Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep 10(10):1681–1691. https://doi.org/10.1016/j.celrep.2015.02.034
Ozkurede U, Miller RA (2019) Improved mitochondrial stress response in long-lived Snell dwarf mice. Aging Cell. https://doi.org/10.1111/acel.13030
Molenaars M, Janssens GE, Williams EG, Jongejan A, Lan J, Rabot S et al (2020) A conserved mito-cytosolic translational balance links two longevity pathways. Cell Metab 31(3):549-563. https://doi.org/10.1016/j.cmet.2020.01.011 (e7)
Funding
This work was supported by FAEPEX, the National Council for Scientific and Technological Development (CNPq; case numbers 303571/2018–7; 140285/2016–4; 442542/2014–3 and 306535/2017–3), the Coordination for the Improvement of Higher Education Personnel (CAPES; finance code 001), and the São Paulo Research Foundation (FAPESP; case numbers 2015/26000–2, 2016/18488–8, 2018/20872–6, 2019/11820–5, 2020/13443–1 and 2021/08692–5).
Author information
Authors and Affiliations
Contributions
VRM and JRP wrote the paper and were ultimately responsible for the experiments in this study. VRM, JDB, RCG, ALR, RFLV, SCBRN and GCA. designed and performed the experiments with animals. BMC and RRB performed the mitochondrial function experiments. VRM, MBS, and FMS designed and performed the cell culture experiments. SQB, CDR, and LAV contributed to PET scan experiments. LAV, FMS, LPM, ASRS, ERR, DEC, and JRP contributed to the discussion and laboratory support. All the authors have read, critically revised, and approved this manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors of this study have no conflict of interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
18_2023_4771_MOESM1_ESM.tif
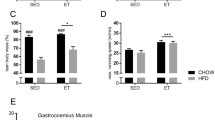
Figure S1. Morphological and physiological parameters. A. Fasting glucose at baseline (before exercise) and after (16 h and 90 h) exercise. B. Fasting insulin at baseline (before exercise) and after (16 h and 90 h) exercise. C. Body weight. D. Lee index. E. Total fat weight. F. Tissue weights (gastrocnemius, heart, mesenteric WAT, retroperitoneal WAT, perigonadal WAT, inguinal WAT, and brown adipose tissue). G. Representative images of hematoxylin–eosin (H&E, 10x, scale bar: 100 µM) in the liver. H. Liver tissue weight. I. Biochemical parameters in the serum (triglycerides, cholesterol, and high-density lipoprotein. J. mRNA levels of gluconeogenic genes (Pepck, G6pase, and Pc). In A-B, n = 7/group was used (* p < 0.05 vs WT group. # p < 0.05 vs Basal condition). In C-F,H n = 9/group was used. In I-J, n = 5/group was used. * p < 0.05 vs WT group. # p < 0.05 vs Slc2a4+/− Sedentary group. (TIF 13534 KB)
18_2023_4771_MOESM2_ESM.tif
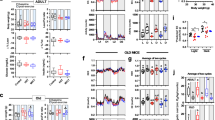
Figure S2. Gene ontology analyses of up/downregulated genes in the skeletal muscle of Slc2a4+/− mice. A. Biological process, cellular components, and molecular functions of upregulated genes in the endurance versus sedentary comparison. B. Biological process, cellular components, and molecular functions of downregulated genes in the endurance versus sedentary comparison. C. Biological process, cellular components, and molecular functions of upregulated genes in the strength versus sedentary comparison. D. Biological process, cellular components, and molecular functions of downregulated genes in the strength versus sedentary comparison. E. Confirmation of common downregulated genes in the gastrocnemius of Slc2a4+/− mice (Cdh4, Ctgf1, Gas1, Hey1, Penk, Ptprb, Amd2, Sorbs2). F-G. Confirmation of common upregulated genes in the gastrocnemius of Slc2a4+/− mice (Srxn1, Irx3, Klhl40, Hspa1a, Hspa1b, Hspb1, Ninj1, Hspb7, Ogdha, Lmcd1, Nfic, Bcl2l13, Retsat, Hspa9, Dnaja4, Fads6, Hadha). In E–G, n = 5, 6, 6 was used. # p < 0.05 vs Slc2a4+/− Sedentary group. (TIF 10541 KB)
18_2023_4771_MOESM3_ESM.tif
Figure S3. Behavioral response after exercise protocols. A. Schematic figure of the groups submitted to the elevated plus maze and open field test (Slc2a4+/− Sedentary, and Slc2a4+/− Endurance or Strength after exercise protocol). B. Time in the border during the open field test. C. Time in the center during the open field test. D. Grooming times during the open field test. E. Percentage of time spent in the open arms during the elevated plus maze test. F. Percentage of time spent in the closed arms during the elevated plus maze test. G. Percentage of time spent in the center during the elevated plus maze test. H. Percentage of entries in the open arms during the elevated plus maze test. I. Corticosterone levels in the serum. In B-H, n = 5/group was used. In I, n = 4, 5, 5 was used. (TIF 3995 KB)
18_2023_4771_MOESM4_ESM.tif
Figure S4. Adipose tissue alterations among the groups. A. Browning-related genes (Pgc1a, Prdm16, Ucp1, Nrf1, and Cs) in the pgWAT. B. Browning-related genes (Pgc1a, Prdm16, Ucp1, Nrf1, Cs) in the ingWAT. C. Browning-related genes (Pgc1a, Prdm16, Ucp1, Nrf1, Cs) in the BAT. D. Representative images of hematoxylin–eosin (H&E, 10x) in the pgWAT. E. Representative images of hematoxylin–eosin (H&E, 10x) in the ingWAT. F. Representative images of hematoxylin–eosin (H&E, 10x, scale bar: 100 µM) in the BAT. In A-C, n = 5/group was used. # p < 0.05 vs Slc2a4+/− Sedentary group. (TIF 20533 KB)
18_2023_4771_MOESM5_ESM.tif
Figure S5. Indirect calorimetry among the groups. A. VO2 consumption during dark and light cycles, and Area under the curve (AUC). B. Heat production during dark and light cycle, and Area under the curve (AUC). C. VCO2 consumption during dark and light cycle, and Area under the curve (AUC). D. Accumulative food intake during dark and light cycle, and Area under the curve (AUC). E. Respiratory exchange ratio (RER) during dark and light cycle, and Area under the curve (AUC). F. Locomotor activity during dark and light cycle, and Area under the curve (AUC). In A-F, n = 5/group was used. * p < 0.05 vs WT group. # p < 0.05 vs Slc2a4+/− Sedentary group. (TIF 9250 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Muñoz, V.R., Botezelli, J.D., Gaspar, R.C. et al. Effects of short-term endurance and strength exercise in the molecular regulation of skeletal muscle in hyperinsulinemic and hyperglycemic Slc2a4+/− mice. Cell. Mol. Life Sci. 80, 122 (2023). https://doi.org/10.1007/s00018-023-04771-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04771-2