Abstract
Targeting airway goblet cell metaplasia is a novel strategy that can potentially reduce the chronic obstructive pulmonary disease (COPD) symptoms. Tumor suppressor liver kinase B1 (LKB1) is an important regulator of the proliferation and differentiation of stem/progenitor cells. In this study, we report that LKB1 expression was downregulated in the lungs of patients with COPD and in those of cigarette smoke-exposed mice. Nkx2.1Cre; Lkb1f/f mice with conditional loss of Lkb1 in mouse lung epithelium displayed airway mucus hypersecretion and pulmonary macrophage infiltration. Single-cell transcriptomic analysis of the lung tissues from Nkx2.1Cre; Lkb1f/f mice further revealed that airway goblet cell differentiation was altered in the absence of LKB1. An organoid culture study demonstrated that Lkb1 deficiency in mouse airway (club) progenitor cells promoted the expression of FIZZ1/RELM-α, which drove airway goblet cell differentiation and pulmonary macrophage recruitment. Additionally, monocyte-derived macrophages in the lungs of Nkx2.1Cre; Lkb1f/f mice exhibited an alternatively activated M2 phenotype, while expressing RELM-α, which subsequently aggravated airway goblet cell metaplasia. Our findings suggest that the LKB1-mediated crosstalk between airway progenitor cells and macrophages regulates airway goblet cell metaplasia. Moreover, our data suggest that LKB1 agonists might serve as a potential therapeutic option to treat respiratory disorders associated with goblet cell metaplasia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Airway mucus is an important part of the fundamental host defense system that enables the clearing of inhaled particles by trapping pathogens to preserve airway sterility and homeostasis. However, goblet cell metaplasia-associated excessive airway mucus is one of the most common symptoms of several lung diseases, including asthma, cystic fibrosis, and chronic obstructive pulmonary disease (COPD), leading to severe clinical outcomes in patients [1,2,3]. Hence, goblet cell fate determination is meticulously regulated during both lung development and postnatal lung epithelial homeostasis, as maintaining an adequate number of mucus-producing goblet cells is not only important for securing a basal mucus secretion in the airways, but also for preventing the occurrence of mucus plugging in the bronchioles [4, 5]. Goblet cells are mainly derived from secretoglobin family 1A member 1 (Scgb1a1, also called as CCSP)-expressing club (previously called Clara) cells and are believed to be tissue-resident facultative epithelial progenitors under the control of physiological or injury-associated signals [6, 7]. The transcription factor SAM-pointed domain containing Ets transcription factor (SPDEF) activates forkhead box A3 (Foxa3) to induce the differentiation of club cells into goblet cells [8]. Foxp1/4 restricts goblet cell differentiation during lung development and regeneration by regulating the anterior gradient 2 (Agr2) [4]. Notch signaling interacts with other pathways through downstream elements, including epidermal growth factor receptor, to direct goblet cell differentiation in the airway epithelium [9,10,11,12]. The club cell-specific activation of β-catenin at the later stages of lung development leads to goblet cell metaplasia, which is associated with an increased secretion of mucins [13]. The epithelial-specific deletion of receptor-like tyrosine kinase (Ryk) gradually increases the number of goblet cells during lung development, while the club cell-specific deletion of Ryk in adult stages also drives goblet cell metaplasia and mucus hypersecretion during regeneration [5]. In addition, external interleukin 33 (IL-33) induces goblet cell differentiation in the intestine and lungs through IL-13, which is usually produced by type-2 innate lymphoid cells [14, 15]. The number and activity of goblet cells is induced by various inflammatory stimuli. However, the regulation of goblet cell differentiation by inflammatory cells remains largely unexplored.
Resistin-like molecule-α (RELM-α, also known as HIMF or FIZZ1), which is a protein produced and secreted by alternatively activated macrophages, epithelial cells, eosinophils, and microglial cells, is highly induced by IL-13 [16,17,18]. RELM-α expression is significantly elevated in the lungs during airway inflammation [19, 20]. The intranasal Alternaria administration-induced expression of RELM-α results in airway eosinophilia, epithelial thickening, and fibrosis [21]. In addition, RELM-α promotes the production of monocyte chemotactic protein-1 (MCP-1) by lung endothelial cells [22]. The transgenic expression of RELM-α in club cells has been shown to increase the abundance of macrophages in bronchoalveolar lavage during silica-induced inflammation [23]. However, the crosstalk between epithelial cells and macrophages in goblet cell fate specification remains largely unknown.
Liver kinase B1 (LKB1), which is encoded by serine/threonine kinase 11 (Stk11), is a tumor suppressor and its inactivation has been demonstrated to drive the development of lung cancer, melanoma, prostate cancer, and cervical cancer [24,25,26]. The loss of LKB1 results in altered energy metabolism, impaired integrity, and disrupted cell polarity in mammalian epithelial tissues [27,28,29,30]. LKB1 deficiency has been demonstrated to impair ciliated cell differentiation in the fallopian tube and lung epithelium [31, 32]. In this study, we employed single-cell RNA sequencing (scRNA-seq) analysis, organoid cultures, transgenic tools, and immunostaining techniques to investigate the role of LKB1 in lung goblet cell fate specification. We found that LKB1 was expressed in club cells, and that adult mice with LKB1 deleted in the embryonic lung epithelium displayed goblet cell metaplasia in the lungs. Furthermore, silencing the epithelial Lkb1 increased the expression of RELM-α, which drives goblet cell differentiation, macrophage accumulation, and activation in the lungs. Thus, macrophage-derived RELM-α contributes to the further development of airway goblet cell metaplasia. This study provides a novel insight into the crosstalk between macrophages and club cells in goblet cell lineage commitment in adult airways.
Materials and methods
Patient tissue samples
The COPD human lung specimens were bronchoscopically obtained from patients with COPD. The normal human lung specimens were obtained from the para-carcinoma tissue of patients with lung cancer during operation. The study was performed in accordance with the 1964 Declaration of Helsinki. All the human lung specimens were obtained from the Research Biorepository under the approval of the Research Ethics Committee of Haihe Hospital, Tianjin University (2021HHKT-018).
Mice
NK2 homeobox 1 (Nkx2.1)-Cre transgenic mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA). Lkb1f/f mice were generously gifted by Dr. Ronald DePinho (Boston, MA, USA). Scgb1a1CreER mice were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). C57BL/6J mice were purchased from Beijing Huafukang Bioscience Co., Ltd. (Beijing, People’s Republic of China). To induce Lkb1 knockdown in the mouse lung epithelium, Nkx2.1Cre; Lkb1f/f mice were established by crossing Nkx2.1-Cre mice with Lkb1f/f mice. Scgb1a1CreER; Lkb1f/f mice were established by crossing Scgb1a1CreER mice with Lkb1f/f mice. Tamoxifen was injected intraperitoneally at 200 mg/kg every other day for three times to induce the deletion of Lkb1 in club cells. Mice were maintained in a specific pathogen-free facility at the Tianjin University Haihe Hospital. All animal studies were approved by the Animal Care and Use Committee of the Tianjin University Haihe Hospital.
Cell culture
The mouse lung fibroblast cell line MLg 2908 (MLg) was purchased from the American Type Culture Collection (Manassas, VA, USA). MLg cells were cultured in DMEM supplemented with 10% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin in a humidified incubator at 37 °C and 5% CO2.
Fluorescence-activated cell sorting
Mouse lungs were dissociated via elastase digestion, as described previously [6]. Briefly, the lungs were perfused with 4 U/mL elastase (Worthington Biochemical Corporation, Lakewood, NJ) and finely minced in a dish containing DNase I (Sigma-Aldrich, St. Louis, MO). The single-cell suspensions prepared for fluorescence-activated cell sorting (FACS) were then passed through a 100 μm strainer (Falcon; BD Biosciences, San Jose, CA) and collected in a 50 mL tube, and RBC lysis buffer (eBioscience, San Diego, CA, USA) was added to the tube. Cells were resuspended in Hanks’ balanced salt solution supplemented with 2% fetal bovine serum (FBS), 10 mM HEPES, 100 mg/mL streptomycin, 100 IU/mL penicillin, and 0.1 mM EDTA. Flow cytometry was performed to sort the club cells from the mouse lung tissues using primary antibodies, including anti-CD31-biotin, anti-CD34-biotin, anti-CD45-biotin, anti-CD24-phycoerythrin (PE), anti-EpCAM-PE-Cyanine 7 (PE-Cy7), Sca-1-allophycocyanin (APC), and the secondary antibody streptavidin APC-eFluor 780. The sorting of macrophages from the mouse lung tissues was performed using APC-Cy7-conjugated anti-mouse CD11b antibody. The dead cells were stained with 7-amino-actinomycin D (7-AAD). All antibodies were obtained from eBioscience (San Diego, CA, USA). Fluorescence-activated cell sorting (FACS) was performed using a FACS Aria III sorter (BD Biosciences, San Jose, CA, USA).
3D organoid culture
As described previously [6], FACS-sorted mouse EpCAM+ CD24+ Sca-1+ airway club cells (3000 cells per well) were co-cultured with MLg lung fibroblast cells in Matrigel. The cell mixture was seeded in the inserts, which were then placed into the wells of a 24-well plate containing basal DMEM culture medium (Cellgro, Manassas, VA) supplemented with 10 μM SB43142. Organoid cultures were maintained at 37 °C in a 5% CO2 incubator, and the culture medium was replaced every other day. Colonies were visualized and counted on day 10 after seeding. In some experiments, club cells were co-cultured with CD11b+ macrophages (1500 cells per well) in the organoids. Organoid cultures were embedded in paraffin for immunofluorescent staining or lysed using TRIzol reagent for gene expression analysis.
Quantitative RT-PCR
The total RNA was extracted from the organoids or lung tissues using TRIzol reagent (Invitrogen) and then reverse-transcribed using M-MLV Reverse Transcriptase (Promega Corporation, Madison, WI, USA). qPCR was conducted using SYBR Select Master Mix (Applied Biosystems, Thermo Fisher Scientific Corporation, Foster City, CA, USA). The relative expression levels of the target genes were normalized to that of the housekeeping gene, Actb, for the same sample. The primer sequences designed for this study are as follows: Muc5AC-F, 5′-TGACTCAATCTGCGTGCCTT-3′; Muc5Ac-R, 5′-AGGCCTTCTTTTGGCAGGTT-3′; CLCA3-F, 5′-GGCATCGTCATCGCCATAG-3′; CLCA3-R, 5′-CACCATGTCCTTTATGTGTTGAATG-3′; Foxa3-F, 5′-CTTGGTGGAGGTTGGGTGAG-3′; Foxa3-R, 5′-ACAGGCAGTATTCCCAAGCC-3′; Spdef-F, 5′-GACTGTGGAATTCCTGGGGG-3′; Spdef-R, 5′-ATTGTGGCAGGAGCAGAGAC-3′; β-actin-F, 5′-GGCCAACCGTGAAAAGATGA-3′; β-actin-R, 5′-CAGCCTGGATGGCTACGTACA-3′.
Hematoxylin and eosin staining
The rehydrated sections were stained with hematoxylin (ZSGB-BIO, Beijing, People’s Republic of China) for 3 min, rinsed in water for 10 min, and dehydrated in 85% and 95% alcohol for 2 min each. They were subsequently stained with an eosin solution (ZSQB-BIO) for 30 s, followed by dehydration in 95% and 100% alcohol for 3 min each, and cleared in xylene twice for 5 min each. The slides were then mounted using a neutral balsam.
Alcian blue staining
Alcian blue staining was performed to identify the goblet cells. The rehydrated sections were stained with 1% Alcian blue (Poly Scientific R&D Corp., Bay Shore, NY, USA) for 10 min and rinsed in running water for 10 min. They were subsequently stained with an eosin solution (ZSQB-BIO) for 30 s, dehydrated, and mounted using a neutral balsam.
Immunofluorescent microscopy
The paraffin-embedded sections (5 μm) of mouse lungs or organoids were subjected to antigen retrieval in citric acid (10 mM, pH 6), blocked with 5% bovine serum albumin in 0.2% Triton-X/PBS, and then stained with the primary antibodies at the following dilutions: rabbit anti-ClCa3 (1:200, Invitrogen), mouse anti-CYP2F2 (1:100, Santa Cruz Biotechnology, Inc., Dallas, TX, USA), rabbit anti-YM1/2 (1:200, Abcam, Cambridge, UK), and rabbit anti-RELM-α (1:200, Abcam). The sections were incubated with Alexa fluorophore-conjugated secondary antibodies (1:200, Invitrogen) at a 1:200 dilution. Then, they were mounted using Fluoromount G containing DAPI and imaged using an IX73 inverted fluorescence microscope (Olympus, Tokyo, Japan) or a laser confocal microscope (Leica TCS SP8). We also used a confocal microscope (DMi8 CS Bino, Leica) to collect fluorescent images using a 10 × objective (Leica, HC PL APO 10 × /0.45), a HIVIS scan optics module with rotation, and an electron-multiplying charge-coupled device camera (Evolve 512; Photometrics). We added scale bars, and processed the images using LAS X (Leica) and Photoshop (Adobe). For the quantification of images, three or more regions were analyzed in each section.
Transcriptomic dataset retrieval
The transcriptomic data of the lung tissues of patients with COPD (GSE106986) and with airway epithelial brushings of asthma (GSE67472), the lung tissues of cigarette smoke-exposed mice (GSE125521), the lung tissues of house dust mite-exposed mice (GSE71822), and the single-cell RNA-seq data of the airway epithelial cells from patients with cystic fibrosis (GSE150674) were obtained from the series matrix files downloaded from the NCBI Gene Expression Omnibus (GEO) database. These transcriptomic data were normalized based on the Counts per Million function and/or the log2 function before the analysis of LKB1 expression.
Single-cell RNA-seq and data analysis
The total lung cells isolated from three Lkb1f/f or Nkx2.1Cre; Lkb1f/f mice were pooled and processed to prepare scRNA-seq libraries using Chromium Single Cell 3′ Reagent v3 Kits (10 × Genomics, Pleasanton, CA, USA). Single-cell suspensions were loaded onto a chromium single-cell controller instrument (10XGenomics, Pleasanton, CA, USA) to generate single-cell gel beads in emulsions (GEMs). After the generation of the GEMs, reverse transcription, cDNA PCR amplification, and library preparation were performed according to the 10 × Genomics 3′ gene expression protocol. These libraries were pooled and sequenced using the HiSeq NovaSeq platform (Illumina, San Diego, CA, USA) in the 150 bp paired-end mode. Single-cell RNA-seq data preprocessing scRNA-seq analyses resulted in demultiplexed cellular barcodes and the reads were mapped to the mouse genome (version mm10) using the Cell Ranger pipeline (version 3.0.0) provided by 10 × Genomics. Seurat (version 3.5.0) was used for subsequent analysis [33]. To remove the low-quality cells and multiple captures, cells were removed if they expressed fewer than 200 unique genes or 500 UMI and more than 5000 unique genes or 50,000 UMI. In addition to these threshold values, we further removed the cells with more than 20% mitochondrial-derived UMI counts. Then, the QC criteria were applied. To reduce the dimensionality of the scRNA-seq dataset, principal component analysis was performed using the RunPCA function in Seurat. For tSNE projection and clustering analysis, the cells were clustered considering a graph-based clustering approach and were visualized in 2D using tSNE. The marker genes of each cluster were identified using the FindAllMarkers function (test. use = bimod) in Seurat. We used the FindMarkers function in Seurat to identify the differentially expressed genes (DEGs) [33]. A p < 0.05 and a |log2foldchange|> 0.58 were set as the threshold values for a significant differential expression. Gene ontology enrichment analyses of the DEGs were performed using the R software based on a hypergeometric distribution.
Pseudotime analysis
Pseudotime analysis was performed on club and goblet cells using the Monocle2 R package (version 2.18.0). The club and goblet cells on the pseudotime trajectory were derived from Nkx2.1Cre; Lkb1f/f and Lkb1f/f mice.
Gene set variation analysis (GSVA)
We performed pathway analyses of the Kyoto Encyclopedia of Genes and Genomes (KEGG) subset of canonical pathways described in the C2 curated gene sets of the molecular signature database (MSigDBwith) of the GSEABase package (version 1.52.1). The GSVA package (version 1.38.2) was used to evaluate the pathway activity estimates.
Transcription factor (TF) regulatory network analysis
We used single-cell regulatory network inference and clustering (SCENIC; version 1.2.4) to perform TF regulatory network analysis. Using RcisTarget (version 1.10.0), mmp9 TFs were downloaded and used as a reference. The DEGs in the lung epithelial cells between Nkx2.1Cre; Lkb1f/f and Lkb1f/f mice were used to create gene regulatory networks using software GENIE3 (version 1.12.0). Cytoscape (version 3.8.2) was used to visualize the TF regulatory network.
Cell–cell communication analysis
Cell–cell interaction analysis was conducted using the scRNA-seq data and the CellChat R package (version 1.1.0) (www.cellchat.org). We evaluated the average expression of each ligand-receptor pair in club cells, goblet cells, resident macrophages, and monocyte-derived macrophages, and only those with a p value lower than 0.05 were used for predicting the potential cell–cell interactions that were associated with Lkb1. When the ligand or the receptor was undetectable, a cell–cell interaction was considered nonexistent.
Statistical analysis
Data are expressed as the mean ± SEM. The significance of the results was assessed using an unpaired t test between the two groups. Results were considered statistically significant at a p ≤ 0.05.
Results
Patients with COPD exhibit a decreased LKB1 expression in the airway epithelium
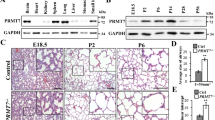
We retrieved the transcriptomic data from 14 patients with COPD and 5 healthy controls from publicly available GEO datasets (GSE106986) [34]. Our analyses revealed that the expression of LKB1 transcripts was significantly reduced in the lung homogenates from patients with COPD than in those from normal controls (Fig. 1a). Reanalysis of the retrieved transcriptomic data from cigarette smoke-exposed mice also showed a decrease in Lkb1 expression in the lungs (GSE125521) (Fig. 1b). Immunohistochemistry staining also showed an evident LKB1 level reduction in the lung epithelium in patients with COPD (Fig. 1c). Consistent with previous findings [35, 36], goblet metaplasia and macrophage accumulation were observed in subjects with COPD (Fig. 1c, d). However, as compared with the controls, LKB1 expression remained unchanged in the airway epithelial cells from patients with asthma or in the lung tissues from house dust mite-exposed mice based on our reanalysis of the retrieved transcriptomic data (GSE67472; GSE71822) (Figure S1A, B). We retrieved and reanalyzed the publicly available scRNA-seq data of airway epithelial cells from patients with cystic fibrosis (GSE150674). We found that compared with normal healthy controls, the LKB1 transcript level remained unchanged in the airway epithelial cells from patients with cystic fibrosis (Figure S2).
LKB1 expression is downregulated in the lungs of patients with COPD. a LKB1 transcript expression in lung homogenates of normal healthy subjects or patients with COPD. b LKB1 transcript expression in lung homogenates of control mice or cigarette smoke-exposed mice. c Immunohistochemistry staining of LKB1 and Alcian blue staining of the lung sections of normal subjects and subjects with COPD. d Immunohistochemistry analysis of CD68 staining of the lung sections of normal subjects and subjects with COPD. Quantification of the CD68-positive cells on the lung sections of normal subjects and subjects with COPD
Lkb1 knockout in lung epithelium promotes goblet cell metaplasia
To investigate whether goblet cell differentiation is regulated by LKB1, we crossed Lkb1f/f mice with Nkx2.1Cre mice to generate Nkx2.1Cre; Lkb1f/f mice, in which LKB1 was specifically deleted in the embryonic and adult lung epithelia. Our findings revealed that the general outlook of Nkx2.1Cre; Lkb1f/f mice was similar to that of Lkb1f/f littermate controls at postnatal day 1 (Figure S3A). At this stage, there was no difference in the body and lung weight between Nkx2.1Cre; Lkb1f/f and Lkb1f/f mice (Figure S3B). However, compared with the controls, airway epithelial thickening was evident in Nkx2.1Cre; Lkb1f/f mice (Figure S3C). This was later demonstrated to be related to an increased number of CCSP-expressing club cells (Figure S3D, E). Goblet cells were not observed in Nkx2.1Cre; Lkb1f/f mice (data not shown). However, we noticed that by postnatal week 2, Nkx2.1Cre; Lkb1f/f mice were smaller and had gained 10–25% less weight than the control littermates (Fig. 2a). By week 5 after birth, the Nkx2.1Cre; Lkb1f/f mice had died (Fig. 2b). Histological examination of the lungs showed massive inflammatory cell infiltration in the peribronchiolar connective tissues, in addition to the thickening of the airway epithelium (Fig. 2c). Compared with the control mice, Alcian blue staining revealed a marked increase in the number of mucous cells within the bronchi of Nkx2.1Cre; Lkb1f/f mice (Fig. 2d). Foxj1 expression was decreased in the absence of LKB1, as revealed by the RT-qPCR analysis of whole lungs (Fig. 2e). However, compared with the controls, the expression of Scgb1a1 and ClCa3 was significantly increased in the lungs of Nkx2.1Cre; Lkb1f/f mice (Fig. 2e). These data suggest that Lkb1 restricts goblet cell differentiation in the lungs.
Epithelial loss of Lkb1 leads to airway goblet cell metaplasia and inflammation. a The body weight of Nkx2.1Cre; Lkb1f/f and Lkb1f/f mice was measured at various time points after birth (n = 5–6 per group). b Survival analysis of Nkx2.1Cre; Lkb1f/f and Lkb1f/f mice after birth (n = 18–23). c Hematoxylin and eosin staining indicated inflammatory infiltration and epithelial alterations in Nkx2.1Cre; Lkb1f/f mice after five postnatal weeks (n = 5). d Alcian blue staining showing airway goblet cell metaplasia in Nkx2.1Cre; Lkb1f/f mice after five postnatal weeks (n = 5). e Quantitative PCR analysis of the indicated genes in the lungs isolated from Nkx2.1Cre; Lkb1f/f and control mice after 5 postnatal weeks (n = 3). Data are representative of two or more independent experiments with the error bars representing the mean ± SD. *p < 0.05
To characterize the mechanisms underlying LKB1-deficiency-induced lung pathology, we performed single-cell RNA-seq analysis of the lungs of Nkx2.1Cre; Lkb1f/f and Lkb1f/f mice. In total, 30,408 lung cells were collected for the scRNA-seq analysis of the whole lung cells isolated from 5-week-old Nkx2.1Cre; Lkb1f/f or Lkb1f/f mice, when the Nkx2.1Cre; Lkb1f/f mice began to die (Fig. 3a, b). We used established markers to validate the cell types in each cluster (Figs. 3a and S4). We employed 2D graphs to reveal the diverse cell lineages, including the epithelium (expressing epithelial cellular adhesion molecule (Epcam)), endothelium (cadherin 5 (Cdh5), Myc target protein 1 (Myct1)), fibroblasts (collagen type I alpha 2 chain (Col1a2), osteoglycin (Ogn)), myeloid cells (triggering receptor expressed on myeloid cells 2 (Trem2)), neutrophils (resistin-like gamma precursor (Retnlg), matrix metallopeptidase 9 (Mmp9)), erythroid-like and erythroid precursor cells (5′-aminolevulinate synthase 2 (Alas2)), lymphoid lineages which separated into B cells (CD19), ILC (interleukin 1 receptor-like 1 (Il1rl1)), natural killer cells (C–C motif chemokine ligand 5 (Ccl5), killer cell lectin-like receptor, family E, member 1 (Klre1)), and T cells (CD3d) (Figure S4). Overall, based on the single-cell map that assigned cell types, the cellular differentiation and transcriptomic profiles of the lungs were further studied.
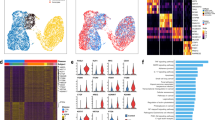
Single-cell RNA-seq analysis demonstrating the stimulated goblet cell differentiation. a, b tSNE plot of 30,408 lung cells pooled from each group (3 mice per group), color-coded by their associated cell lineages a or the origin of the sample type b. c tSNE plot of 3,373 lung epithelial cells, color-coded by their associated cell types. d Enrichment of the gene expression in each lung epithelial type. Node size is proportional to the percentage of cells in the cluster expressing a gene. Node color is proportional to the average expression level for the gene in the cluster. e Network plot revealing the expression of genes that changed between the epithelial cells of Nkx2.1Cre; Lkb1f/f mice and those of Lkb1f/f mice and the genes in the lung disease database (https://www.disgenet.org/home/). The red connecting lines show the upregulated genes and the blue connecting lines show the downregulated genes in the epithelial cells of Nkx2.1Cre; Lkb1f/f mice compared with those of Lkb1f/f mice. The number of connecting lines represents the number of epithelial cell types that express these genes. The color of the gene nodes (from white to dark brown) represents the number of epithelial cell types (from low to high), respectively. The lung disease types are listed in order. The color of the disease number (from white to black) represents the number of genes related to the indicated disease (from low to high), respectively. f Predicted effect of Lkb1 on the differentiation model of club cells into goblet cells, and predicted differentiation lineage model between club and goblet cells
We further used marker genes to enhance the annotation of the epithelial clusters, generating seven epithelial subpopulations, including AT2 cells (expressing known marker ATP-binding cassette subfamily A member 3 (Abca3), surfactant protein C (Sftpc)), AT1 cells (aquaporin 5 (Aqp5), podoplanin (Pdpn)), basal cells (tumor protein 63 (Trp63), keratin 5 (Krt5)), club cells (cytochrome P450 2F2 (Cyp2f2), Scgb1a1, ciliated cells (cadherin-related family member 3 (Cdhr3), forkhead box J1 (Foxj1)), goblet cells (Foxa3, Spdef), and neuroendocrine cells (chromogranin A (Chga), achaete-scute homolog 1 gene (Ascl1)) (Fig. 3c, d).
A comparison of the network plot of the DEGs of lung epithelial cells between Nkx2.1Cre; Lkb1f/f and littermate controls showed that the epithelial-derived LKB1-relevant lung diseases were ranked by lung carcinoma, as expected, since LKB1 is known as a tumor suppressor (Fig. 3e). Among the 30 Nkx2.1Cre; Lkb1f/f mice investigated, we found one in which lung carcinoma was developing (Figure S5). Other relevant diseases included COPD, pulmonary fibrosis, pneumonia, and influenza (Fig. 3e). Pseudotime analysis revealed the differentiation trajectories of club cells to goblet cells and showed that almost all of the goblet cells mapped to the club cell differentiation trajectory were derived from Nkx2.1Cre; Lkb1f/f mice, confirming the inhibitory role of LKB1 in goblet cell differentiation (Fig. 3f).
KEGG pathway signatures and SCENIC analysis suggested that the Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling pathway may be affected by Lkb1 (Fig. 4a, b). The Stat3 mRNA levels were elevated in LKB1-deficient club cells compared to those in normal club cells (Fig. 4b, c). Consistently, immunofluorescence staining verified the upregulation of phosphorylated STAT3 (pSTAT3) in club cells in the absence of LKB1 (Fig. 4d). These data suggest that STAT3 may be involved in club cell fate determination.
Molecular effects of Lkb1 knockdown in club cells. a Discrepancies in pathway activities scored using GSVA, between the club cells from Nkx2.1Cre; Lkb1f/f mice versus those from Lkb1f/f mice. b Transcriptional network revealing the transcription factors (TFs) identified in the epithelium of Nkx2.1Cre; Lkb1f/f mice versus Lkb1f/f mice via SCENIC. The outer nodes represent the upregulated and downregulated TFs (in red and blue, respectively). The color of the TFs (from light to dark) reveals the number of target genes regulated by the TFs (from low to high), respectively. The inner nodes indicate the subpopulations of lung epithelium and the sizes of the nodes represent the number of target genes involved in this subpopulation. c Violin plots of Stat3 expression in the club cells of Nkx2.1Cre; Lkb1f/f mice compared with those of Lkb1f/f mice. d Immunofluorescent staining of the lung sections from Nkx2.1Cre; Lkb1f/f mice versus those from Lkb1f/f mice with phosphorylated STAT3 and Cyp2f2 (n = 3)
Lkb1 silencing upregulates RELM-α in club cells that regulate goblet cell differentiation
To investigate the molecular mechanism of LKB1 in the regulation of club cell differentiation into goblet cells, we compared the transcriptomic profiles of club cells in the scRNA-seq data from Nkx2.1Cre; Lkb1f/f mice and in the littermate controls using scRNA-sequencing. We observed 136 differentially expressed transcripts with fold changes of over 1.5-fold with 113 upregulated and 23 downregulated transcripts. The top upregulated transcripts in the club cells of Nkx2.1Cre; Lkb1f/f mice included resistin-like molecule alpha (Retnla) and chitinase-like protein 3 (Chil3) (Figure S6A, Table S1). The increase in Retnla and Chil3 expression was confirmed via bulk RNA-seq analysis of the lung epithelial cells sorted from Nkx2.1Cre; Lkb1f/f mice and the littermate controls (Figure S6B, Table S2). The transcriptomic profiles identified 120 upregulated and 77 downregulated transcripts in goblet cells in the absence of LKB1 (Figure S6C). The elevated transcripts included Clca1 (79-fold), Agr2 (fivefold), and Muc5b (fivefold), which are related to mucus production or secretion (Table S3). We found that the Retnla expression levels were increased in both club cells and goblet cells in the absence of LKB1, along with the upregulation of mucus-related and inflammation-related genes, such as Muc5b, Chil3, Cd9, and Tnfaip3 (fold change > 1.5; p < 0.05) (Fig. 5a). These data suggest that Retnla may regulate goblet cell differentiation.
Lkb1 knockdown promotes RELM-α, which drives club cell differentiation into goblet cells. a Heatmap revealing the co-upregulated DEGs of club cells and goblet cells which are related to the extracellular region, inflammation, and regulation of macrophages in Nkx2.1Cre; Lkb1f/f mice versus Lkb1f/f mice. b Immunofluorescent staining of the lung sections from Nkx2.1Cre; Lkb1f/f mice versus Lkb1f/f mice with Cyp2f2 and RELM-α or with Muc5Ac and RELM-α (n = 3). c Gene expression analysis of the indicated transcripts using RT-qPCR (relative to β-actin) in the organoid cultures of club cells in the presence or absence of RELM-α (n = 5). Data are representative of two or more independent experiments with the error bars representing the mean ± SD. *p < 0.05
Confocal microscopy revealed that RELM-α, the protein encoded by Retnla, was undetectable in normal mouse club and goblet cells (Fig. 5b). However, RELM-α was found to be upregulated and co-localized with Cyp2F2 (club cell marker) and Muc5Ac (goblet cell marker) in the lungs of Nkx2.1Cre; Lkb1f/f mice (Fig. 5b). Airway goblet cells expressed lower levels of Cyp2F2 than club cells, in Nkx2.1Cre; Lkb1f/f mice (Figure S7). Immunostaining of the adjacent lung sections suggested the presence of two types of goblet cells: one expressing RELM-α and the other expressing little or no RELM-α (Figure S7). These data suggest that LKB1 inhibits the expression of RELM-α in club cells.
To examine whether RELM-α modulates goblet cell differentiation in general, we implemented an in vitro 3D organoid culture established previously [6]. Club cells characterized by a CD31−CD34−CD45−EpCAM+ Sca1+CD24low phenotype were isolated from C57BL/6 mice via FACS, followed by an organoid culture with supportive lung fibroblast cells (Figure S8A, B). The size and forming efficiency of club cell-derived organoids remained unchanged in the presence of recombinant RELM-α (Figure S8B–D), suggesting that RELM-α plays little role in club cell proliferation. However, the expression of the goblet cell markers Muc5Ac, ClCa3, and Foxa3 was promoted in organoid cultures supplemented with recombinant RELM-α based on qRT-PCR analysis (Fig. 5c). To confirm the role of Lkb1 in goblet cell differentiation, we established Scgb1a1CreER; Lkb1f/f mice which exhibited conditional loss of Lkb1 in airway club cells after tamoxifen injection (Figure S9A). As predicted, the organoid cultures of mouse airway club cells indicated that Clca3 gene expression was promoted in absence of Lkb1 (Figure S9B). The results of the protein–protein interaction prediction tool STRING or GeneMANIA analysis showed no evidence of the interaction between Lkb1 and RELM-α (Figure S10, 11). Taken together, these data suggest that RELM-α promotes goblet cell differentiation.
Epithelial silencing of Lkb1 induces macrophage accumulation and activation
RELM-α drives the recruitment of monocytes and promotes the alternative activation of pulmonary macrophages [21, 37,38,39]. To examine whether RELM-α upregulation results in the accumulation and activation of macrophages, we performed immunofluorescent staining of lung sections using antibodies against chitinase-like 3 (Chil3/Ym1), Muc5Ac, and Cyp2F2. We observed that the Ym1-positive macrophages aggregated around the airway Cyp2F2-positive cells (Fig. 6a). Confocal immunofluorescent staining results demonstrated that the majority of the Muc5Ac-positive goblet cells were stained with Ym1, whereas no immunoreactive Ym1 was found in the Cyp2f2-hi club cells of Nkx2.1Cre; Lkb1f/f mice (Figure S12A, B). To examine whether macrophages affect the differentiation of club cells into goblet cells, we isolated CD11b+ macrophages from lung tissues of C57BL/6 mice by FACS and co-cultured them with club cells at a 1:2 ratio (Figs. 6b, S13). Quantitative PCR analysis indicated that the expression of goblet cell markers Clca3, Muc5ac, Spdef was increased in organoid cultures in presence of macrophages (Fig. 6c).
Segregated macrophages around airway club cells in Nkx2.1Cre; Lkb1f/f mice. a Immunofluorescent staining of the lung sections from Nkx2.1Cre; Lkb1f/f mice versus Lkb1f/f mice with Cyp2f2 and YM1 (n = 3). b Schematic illustration of the macrophage-club co-culture organoid model. c Quantitative PCR analysis of the indicated transcripts (relative to β-actin) in the organoid cultures of mouse club cells with or without macrophages (n = 5). d, e tSNE plot of 5051 lung myeloid cells, color-coded by their associated cell lineages d or the origin of the sample type e. f Existing markers identifying the source of the resident or monocyte-derived macrophages in the lungs. g Known markers showing that part of the monocyte-derived macrophages are of an alternatively activated phenotype. Data are representative of two or more independent experiments with the error bars representing the mean ± SD. *p < 0.05
Next, we characterized these macrophages based on single-cell RNA-seq data analysis with marker genes (Figure S14). Lung myeloid cells can be segregated into three distinct cell types: dendritic cells (interferon regulatory factor 8 [Irf8], C-type lectin domain containing 9A [Clec9a]), monocytes (adhesion G protein-coupled receptor E4 (Adgre4)), or macrophages (mannose receptor C-type 1 (Mrc1), heme oxygenase 1 (Hmox1)) (Figs. 6d, S14). Consistent with the findings mentioned above (Fig. 6a), the macrophage population was more abundant in the lungs of Nkx2.1Cre; Lkb1f/f mice than in those of control mice (Fig. 6e). To explore the origin of these macrophages, we used established cell markers that have been reported as tissue-resident or monocyte-derived macrophage markers to segregate the macrophage clusters [40,41,42]. Lung resident macrophages were identified as clusters expressing higher levels of sialic acid-binding Ig-like lectin F (Siglecf), macrophage receptor with collagenous structure (Marco), and peroxisome proliferator-activated receptor-gamma (Pparg), and monocyte-derived macrophages were identified as clusters expressing high levels of MAF BZIP TF B (Mafb), integrin subunit alpha M (Itgam), and apolipoprotein E (Apoe) (Fig. 6f). Some monocyte-derived macrophages expressed Retnla, Arg1, and Chil3, which is indicative of their alternatively activated M2 phenotype (Fig. 6g). M2 macrophages were observed in Nkx2.1Cre; Lkb1f/f mice, but not in control mice (Fig. 6e). Collectively, these data suggest that the suppression of epithelial LKB1 induces the recruitment of monocytes to the lungs and promotes their M2 phenotype activation.
To further understand how the origin of macrophages affects club cell differentiation in the absence of Lkb1, we analyzed the cell–cell interactions between macrophages and club cells, as well as between macrophages and goblet cells, and found that club cells interact with monocyte-derived macrophages in different ways from those with the resident macrophages (Fig. 7a). For instance, monocyte-derived macrophages have a greater impact on the differentiation of mouse club cells (Fig. 7a). Altogether, these data suggest that LKB1 is an inhibitory factor in regulating airway goblet cell differentiation. Lkb1 loss may upregulate the airway progenitor cell expression of RELM-α, which promotes airway goblet cell differentiation and pulmonary macrophage infiltration (Fig. 7b). By secreting RELM-α, monocyte-derived macrophages differentiate into the M2 phenotype to further enhance goblet cell fate.
Lkb1 mediated cell–cell interactions among club cells, goblet cells, and macrophages. a Gene ontology enrichment analyses based on the differences in the receptor and ligand networks among club cells, goblet cells, and macrophages. b Schematic expression of the LKB1-mediated crosstalk between epithelial progenitor cells and macrophages in goblet cell differentiation
Discussion
In the present study, we observed that LKB1 restricts goblet cell differentiation in mouse airways. Under normal conditions, the presence of goblet cells in mouse airways is not evident. However, when LKB1 expression is inhibited from the embryonic stage, goblet cell metaplasia develops in the adult stage. In the absence of LKB1, the level of monocyte attracting factor RELM-α is increased in club cells and promotes goblet cell differentiation. Macrophages are also found to be in close proximity to goblet cell metaplasia. ScRNA-seq analysis also indicated the monocytic origin of most infiltrated macrophages. Our data indicate that monocyte-derived macrophages interact with club cells and contribute to goblet cell differentiation and monocyte recruitment to the lungs. We observed a downregulation of LKB1 and macrophage accumulation in subjects with COPD, suggesting a possible role of LKB1 in goblet cell metaplasia in COPD pathogenesis.
LKB1 is widely expressed in human adult tissues and serves as a suppressor of neoplastic transformation. Deletion, mutation, or a reduced expression of LKB1 has been demonstrated to increase the risk of malignant tumors in lungs, colon, breast, cervical, gastrointestinal, and pancreatic tissues through AMP-activated protein kinase (AMPK)-mediated metabolic reprogramming [43]. LKB1 maintains stem cell quiescence and survival, restricting the entry of stem cells into the cell cycle [44, 45]. Airway branching and normal ciliated cell differentiation require LKB1 [32, 46]. However, Lkb1 represses the differentiation of intestinal stem cells into secretory lineages and mucin production by secretory cells [47, 48]. Similar to the intestine, we observed that the silencing of epithelial Lkb1 causes goblet cell metaplasia and mucus overproduction through RELM-α in the lungs. RELM-α plays a role in the recruitment of monocytes/macrophages [21, 37, 49]. An uncontrolled goblet cell differentiation is a prominent feature of a number of respiratory diseases, including COPD and asthma. Mucin hypersecretion prevents mucociliary clearance and reduces gas exchange. We found that the LKB1 transcript levels were lower in the airway epithelium of patients with COPD than in healthy subjects. None of the existing medicines for COPD, including bronchodilators and anti-inflammatory steroids, targets goblet cell differentiation and mucus secretion [50]. Therefore, these medicines suppress COPD symptoms without treating the condition. Therefore, restoring LKB1 expression in airway progenitor cells is a potential therapeutic strategy to tackle COPD by limiting goblet cell differentiation and mucus hypersecretion. It is important to investigate whether the existing drugs for COPD impair LKB1 expression in airway epithelial progenitor cells.
In addition to the direct role of LKB1 in limiting goblet cell differentiation, it indirectly controls goblet cell differentiation by affecting the crosstalk between club cells and macrophages. Club-macrophage interactions have been suggestive but the underlying mechanisms remain largely unknown. The lack of CCSP has been associated with COPD development [51], and recombinant rat CCSP treatment was shown to lead to a macrophage number decrease [52]. The recruitment and activation of macrophages are normally observed in the lungs of patients with COPD [53]. A previous report demonstrated that the inhibition of Lkb1 promotes the recruitment of monocytes and macrophages in the kidneys [54]. We observed that monocyte-derived macrophages were adjacent to airway progenitor cells in the absence of LKB1 in the lungs. These macrophages are monocyte-derived and display an alternative M2 phenotype with RELM-α expression, that favors the differentiation of club cells into goblet cells. Moreover, M2 macrophages are also prominent in the lungs of patients with COPD [55]. Our data suggest that M2 macrophages direct airway epithelial progenitor cell fate by detouring to the goblet cell phenotype. The elimination of lung M2 macrophages may improve goblet cell metaplasia in COPD. However, the resident macrophages play different roles in airway epithelial stem/progenitor cell fate specification since they interact with club cells in different ways from those with monocyte-derived macrophages. Dagher et al. showed that IL33 in resident M2 macrophages favors club cell regeneration [56]. Additionally, it is also known to induce goblet cell differentiation [57, 58].
RELM-α has been reported to be upregulated in the epithelial cells of asthmatic mouse lungs [59], in which goblet cell metaplasia occurs persistently. The increase in the RELM-α level in airway epithelial cells in the Alternaria-induced asthmatic mouse model was STAT6-dependent [21], while that in the oncostatin M-induced mouse model did not require STAT6 [39]. These studies suggest that RELM-α affects airway goblet cell differentiation through multiple pathways. RELM-α is regulated by STAT6 in macrophages [60]. However, it remains unknown whether STAT3 modulates RELM-α expression. STAT3 was previously shown to be associated with the expression of mucus-associated molecules in the intestine [61]. In this study, the hallmark pathway gene signature and SCENIC predicted STAT3 to be a TF responsible for goblet cell hyperplasia in the lungs. We detected an increased phosphorylated STAT3 level, in line with recent studies, confirming that an increased airway goblet cell hyperplasia may be accompanied by an elevated activation of STAT3 [62, 63].
This study has several limitations. First, airway goblet hyperplasia is also a feature of asthma, but our analysis of the retrieved transcriptomic data indicated that LKB1 expression was not altered in the lungs of asthma patients or cigarette smoke-exposed mice. It remains unknown whether a LKB1 downregulation is associated with asthma/COPD overlap pathology. Second, Nkx2.1Cre; LKB1f/f mice started to develop airway goblet cell hyperplasia at 2 weeks and died at the age of 5 weeks (equivalent to 30 years of human age), while COPD commonly occurs in people over 40 years old. It is not known how age impacts Lkb1 expression in healthy subjects or in patients with COPD. Third, the activation of AMP-activated kinase, a downstream target of LKB1, was shown to reduce excessive airway inflammation that may be beneficial to patients with cystic fibrosis. It would be worthwhile to explore whether LKB1 is developmentally defective in this genetic disease in the future since we were unable to. Fourth, our protein–protein interaction prediction results suggest that Lkb1 does not interact with RELM-α directly. Further functional experiments must be done to verify this. The sources of RELM-α were from both LKB1-deficient airway epithelial cells and M2 macrophages surrounding the epithelial cells, but the underlying mechanism requires further investigation. Future investigations on the immunological regulation of the lung epithelial stem/progenitor cell fate, especially the crosstalk between them and macrophages or other immune cells, will provide insights into the pathogenesis of COPD and the development of new therapeutic targets.
Availability of data and material
All data and material used in the study are available.
Code availability
Not applicable.
Abbreviations
- LKB1:
-
Liver kinase B1
- COPD:
-
Chronic obstructive pulmonary disease
- AT2:
-
Alveolar epithelial type II cells
- AT1:
-
Alveolar epithelial type I cells
- ILC:
-
Innate lymphoid cell
- DC:
-
Dendritic cell
- NK:
-
Natural killer
- ScRNA-seq:
-
Single-cell RNA sequencing
References
Shaykhiev R (2019) Emerging biology of persistent mucous cell hyperplasia in COPD. Thorax 74:4–6. https://doi.org/10.1136/thoraxjnl-2018-212271
Kuyper LM, Pare PD, Hogg JC, Lambert RK, Ionescu D, Woods R, Bai TR (2003) Characterization of airway plugging in fatal asthma. Am J Med 115:6–11. https://doi.org/10.1016/s0002-9343(03)00241-9
Gehrig S, Duerr J, Weitnauer M, Wagner CJ, Graeber SY, Schatterny J, Hirtz S, Belaaouaj A, Dalpke AH, Schultz C et al (2014) Lack of neutrophil elastase reduces inflammation, mucus hypersecretion, and emphysema, but not mucus obstruction, in mice with cystic fibrosis-like lung disease. Am J Respir Crit Care Med 189:1082–1092. https://doi.org/10.1164/rccm.201311-1932OC
Li S, Wang Y, Zhang Y, Lu MM, DeMayo FJ, Dekker JD, Tucker PW, Morrisey EE (2012) Foxp1/4 control epithelial cell fate during lung development and regeneration through regulation of anterior gradient 2. Development 139:2500–2509. https://doi.org/10.1242/dev.079699
Kim HT, Yin W, Nakamichi Y, Panza P, Grohmann B, Buettner C, Guenther S, Ruppert C, Kobayashi Y, Guenther A et al (2019) WNT/RYK signaling restricts goblet cell differentiation during lung development and repair. Proc Natl Acad Sci USA 116:25697–25706. https://doi.org/10.1073/pnas.1911071116
Chen H, Matsumoto K, Brockway BL, Rackley CR, Liang J, Lee JH, Jiang D, Noble PW, Randell SH, Kim CF et al (2012) Airway epithelial progenitors are region specific and show differential responses to bleomycin-induced lung injury. Stem Cells 30:1948–1960. https://doi.org/10.1002/stem.1150
Li K, Li M, Li W, Yu H, Sun X, Zhang Q, Li Y, Li X, Li Y, Abel ED et al (2019) Airway epithelial regeneration requires autophagy and glucose metabolism. Cell Death Dis 10:875. https://doi.org/10.1038/s41419-019-2111-2
Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, Gregorieff A, Clevers H, Whitsett JA (2009) SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J Clin Investig 119:2914–2924. https://doi.org/10.1172/JCI39731
Tsao PN, Wei SC, Wu MF, Huang MT, Lin HY, Lee MC, Lin KM, Wang IJ, Kaartinen V, Yang LT et al (2011) Notch signaling prevents mucous metaplasia in mouse conducting airways during postnatal development. Development 138:3533–3543. https://doi.org/10.1242/dev.063727
Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, Rajagopal J (2009) Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 136:1751–1759. https://doi.org/10.1242/dev.029249
Lafkas D, Shelton A, Chiu C, de Leon BG, Chen Y, Stawicki SS, Siltanen C, Reichelt M, Zhou M, Wu X et al (2015) Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature 528:127–131. https://doi.org/10.1038/nature15715
Kang JH, Lee EH, Park SW, Chung IY (2011) MUC5AC expression through bidirectional communication of Notch and epidermal growth factor receptor pathways. J Immunol 187:222–229. https://doi.org/10.4049/jimmunol.1003606
Mucenski ML, Nation JM, Thitoff AR, Besnard V, Xu Y, Wert SE, Harada N, Taketo MM, Stahlman MT, Whitsett JA (2005) Beta-catenin regulates differentiation of respiratory epithelial cells in vivo. Am J Physiol Lung Cell Mol Physiol 289:L971-979. https://doi.org/10.1152/ajplung.00172.2005
Waddell A, Vallance JE, Hummel A, Alenghat T, Rosen MJ (2019) IL-33 induces murine intestinal goblet cell differentiation indirectly via innate lymphoid cell IL-13 secretion. J Immunol 202:598–607. https://doi.org/10.4049/jimmunol.1800292
Ito R, Maruoka S, Soda K, Katano I, Kawai K, Yagoto M, Hanazawa A, Takahashi T, Ogura T, Goto M et al (2018) A humanized mouse model to study asthmatic airway inflammation via the human IL-33/IL-13 axis. JCI Insight. https://doi.org/10.1172/jci.insight.121580
Littlefield A, Kohman RA (2017) Differential response to intrahippocampal interleukin-4/interleukin-13 in aged and exercise mice. Neuroscience 343:106–114. https://doi.org/10.1016/j.neuroscience.2016.11.027
Raes G, De Baetselier P, Noel W, Beschin A, Brombacher F, Hassanzadeh GhG (2002) Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J Leukoc Biol 71:597–602
Pesce JT, Ramalingam TR, Wilson MS, Mentink-Kane MM, Thompson RW, Cheever AW, Urban JF Jr, Wynn TA (2009) Retnla (relmalpha/fizz1) suppresses helminth-induced Th2-type immunity. PLoS Pathog 5:e1000393. https://doi.org/10.1371/journal.ppat.1000393
Zhao J, Jiao X, Wu J, Wang J, Gong W, Liu F, Liu W, Bi W, Dong L (2015) FIZZ1 promotes airway remodeling in asthma through the PTEN signaling pathway. Inflammation 38:1464–1472. https://doi.org/10.1007/s10753-015-0121-5
Wang J, Li F, Yang M, Wu J, Zhao J, Gong W, Liu W, Bi W, Dong L (2014) FIZZ1 promotes airway remodeling through the PI3K/Akt signaling pathway in asthma. Exp Ther Med 7:1265–1270. https://doi.org/10.3892/etm.2014.1580
Doherty TA, Khorram N, Sugimoto K, Sheppard D, Rosenthal P, Cho JY, Pham A, Miller M, Croft M, Broide DH (2012) Alternaria induces STAT6-dependent acute airway eosinophilia and epithelial FIZZ1 expression that promotes airway fibrosis and epithelial thickness. J Immunol 188:2622–2629. https://doi.org/10.4049/jimmunol.1101632
Yamaji-Kegan K, Su Q, Angelini DJ, Champion HC, Johns RA (2006) Hypoxia-induced mitogenic factor has proangiogenic and proinflammatory effects in the lung via VEGF and VEGF receptor-2. Am J Physiol Lung Cell Mol Physiol 291:L1159-1168. https://doi.org/10.1152/ajplung.00168.2006
Madala SK, Edukulla R, Davis KR, Schmidt S, Davidson C, Kitzmiller JA, Hardie WD, Korfhagen TR (2012) Resistin-like molecule alpha1 (Fizz1) recruits lung dendritic cells without causing pulmonary fibrosis. Respir Res 13:51. https://doi.org/10.1186/1465-9921-13-51
Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA et al (2007) LKB1 modulates lung cancer differentiation and metastasis. Nature 448:807–810. https://doi.org/10.1038/nature06030
Hermanova I, Zuniga-Garcia P, Caro-Maldonado A, Fernandez-Ruiz S, Salvador F, Martin-Martin N, Zabala-Letona A, Nunez-Olle M, Torrano V, Camacho L et al (2020) Genetic manipulation of LKB1 elicits lethal metastatic prostate cancer. J Exp Med. https://doi.org/10.1084/jem.20191787
Zhou W, Zhang J, Marcus AI (2014) LKB1 tumor suppressor: therapeutic opportunities knock when LKB1 is inactivated. Genes Dis 1:64–74. https://doi.org/10.1016/j.gendis.2014.06.002
Kottakis F, Nicolay BN, Roumane A, Karnik R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T et al (2016) LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature 539:390–395. https://doi.org/10.1038/nature20132
Han SH, Malaga-Dieguez L, Chinga F, Kang HM, Tao J, Reidy K, Susztak K (2016) Deletion of Lkb1 in renal tubular epithelial cells leads to CKD by altering metabolism. J Am Soc Nephrol 27:439–453. https://doi.org/10.1681/ASN.2014121181
Krawchuk D, Anani S, Honma-Yamanaka N, Polito S, Shafik M, Yamanaka Y (2015) Loss of LKB1 leads to impaired epithelial integrity and cell extrusion in the early mouse embryo. J Cell Sci 128:1011–1022. https://doi.org/10.1242/jcs.162156
Li J, Liu J, Li P, Mao X, Li W, Yang J, Liu P (2014) Loss of LKB1 disrupts breast epithelial cell polarity and promotes breast cancer metastasis and invasion. J Exp Clin Cancer Res 33:70. https://doi.org/10.1186/s13046-014-0070-0
George SH, Milea A, Sowamber R, Chehade R, Tone A, Shaw PA (2016) Loss of LKB1 and p53 synergizes to alter fallopian tube epithelial phenotype and high-grade serous tumorigenesis. Oncogene 35:59–68. https://doi.org/10.1038/onc.2015.62
Chu Q, Yao C, Qi X, Stripp BR, Tang N (2019) STK11 is required for the normal program of ciliated cell differentiation in airways. Cell Discov 5:36. https://doi.org/10.1038/s41421-019-0104-z
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420. https://doi.org/10.1038/nbt.4096
Heinbockel L, Marwitz S, Schromm AB, Watz H, Kugler C, Ammerpohl O, Schnepf K, Rabe KF, Droemann D, Goldmann T (2018) Identification of novel target genes in human lung tissue involved in chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis 13:2255–2259. https://doi.org/10.2147/COPD.S161958
Wang Y, Xu J, Meng Y, Adcock IM, Yao X (2018) Role of inflammatory cells in airway remodeling in COPD. Int J Chron Obstruct Pulmon Dis 13:3341–3348. https://doi.org/10.2147/COPD.S176122
Kim V, Criner GJ (2013) Chronic bronchitis and chronic obstructive pulmonary disease. Am J Respir Crit Care Med 187:228–237. https://doi.org/10.1164/rccm.201210-1843CI
Liu T, Yu H, Ullenbruch M, Jin H, Ito T, Wu Z, Liu J, Phan SH (2014) The in vivo fibrotic role of FIZZ1 in pulmonary fibrosis. PLoS One 9:e88362. https://doi.org/10.1371/journal.pone.0088362
Munitz A, Waddell A, Seidu L, Cole ET, Ahrens R, Hogan SP, Rothenberg ME (2008) Resistin-like molecule alpha enhances myeloid cell activation and promotes colitis. J Allergy Clin Immunol 122(1200–1207):e1201. https://doi.org/10.1016/j.jaci.2008.10.017
Ho L, Yip A, Lao F, Botelho F, Richards CD (2020) RELMalpha is induced in airway epithelial cells by oncostatin M without requirement of STAT6 or IL-6 in mouse lungs in vivo. Cells. https://doi.org/10.3390/cells9061338
Lee YG, Jeong JJ, Nyenhuis S, Berdyshev E, Chung S, Ranjan R, Karpurapu M, Deng J, Qian F, Kelly EA et al (2015) Recruited alveolar macrophages, in response to airway epithelial-derived monocyte chemoattractant protein 1/CCl2, regulate airway inflammation and remodeling in allergic asthma. Am J Respir Cell Mol Biol 52:772–784. https://doi.org/10.1165/rcmb.2014-0255OC
Reyfman PA, Washko GR, Dransfield MT, Spira A, Han MK, Kalhan R (2018) Defining impaired respiratory health. A paradigm shift for pulmonary medicine. Am J Respir Crit Care Med 198:440–446. https://doi.org/10.1164/rccm.201801-0120PP
Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR et al (2019) Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20:163–172. https://doi.org/10.1038/s41590-018-0276-y
Zhang Y, Meng Q, Sun Q, Xu ZX, Zhou H, Wang Y (2021) LKB1 deficiency-induced metabolic reprogramming in tumorigenesis and non-neoplastic diseases. Mol Metab 44:101131. https://doi.org/10.1016/j.molmet.2020.101131
Gan B, Hu J, Jiang S, Liu Y, Sahin E, Zhuang L, Fletcher-Sananikone E, Colla S, Wang YA, Chin L et al (2010) Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468:701–704. https://doi.org/10.1038/nature09595
Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, Tzatsos A, Ozsolak F, Milos P, Ferrari F et al (2010) The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 468:659–663. https://doi.org/10.1038/nature09572
Lo B, Strasser G, Sagolla M, Austin CD, Junttila M, Mellman I (2012) Lkb1 regulates organogenesis and early oncogenesis along AMPK-dependent and -independent pathways. J Cell Biol 199:1117–1130. https://doi.org/10.1083/jcb.201208080
Gao Y, Yan Y, Tripathi S, Pentinmikko N, Amaral A, Paivinen P, Domenech-Moreno E, Andersson S, Wong IPL, Clevers H et al (2020) LKB1 represses ATOH1 via PDK4 and energy metabolism and regulates intestinal stem cell fate. Gastroenterology 158(1389–1401):e1310. https://doi.org/10.1053/j.gastro.2019.12.033
Shorning BY, Zabkiewicz J, McCarthy A, Pearson HB, Winton DJ, Sansom OJ, Ashworth A, Clarke AR (2009) Lkb1 deficiency alters goblet and paneth cell differentiation in the small intestine. PLoS One 4:e4264. https://doi.org/10.1371/journal.pone.0004264
Yamaji-Kegan K, Su Q, Angelini DJ, Myers AC, Cheadle C, Johns RA (2010) Hypoxia-induced mitogenic factor (HIMF/FIZZ1/RELMalpha) increases lung inflammation and activates pulmonary microvascular endothelial cells via an IL-4-dependent mechanism. J Immunol 185:5539–5548. https://doi.org/10.4049/jimmunol.0904021
Gross NJ, Barnes PJ (2017) New therapies for asthma and chronic obstructive pulmonary disease. Am J Respir Crit Care Med 195:159–166. https://doi.org/10.1164/rccm.201610-2074PP
Van Vyve T, Chanez P, Bernard A, Bousquet J, Godard P, Lauwerijs R, Sibille Y (1995) Protein content in bronchoalveolar lavage fluid of patients with asthma and control subjects. J Allergy Clin Immunol 95:60–68. https://doi.org/10.1016/s0091-6749(95)70153-2
Pang M, Liu HY, Li T, Wang D, Hu XY, Zhang XR, Yu BF, Guo R, Wang HL (2018) Recombinant club cell protein 16 (CC16) ameliorates cigarette smoke induced lung inflammation in a murine disease model of COPD. Mol Med Rep 18:2198–2206. https://doi.org/10.3892/mmr.2018.9216
Matera MG, Cazzola M, Page C (2020) Prospects for COPD treatment. Curr Opin Pharmacol 56:74–84. https://doi.org/10.1016/j.coph.2020.11.003
Viau A, Bienaime F, Lukas K, Todkar AP, Knoll M, Yakulov TA, Hofherr A, Kretz O, Helmstadter M, Reichardt W et al (2018) Cilia-localized LKB1 regulates chemokine signaling, macrophage recruitment, and tissue homeostasis in the kidney. EMBO J. https://doi.org/10.15252/embj.201798615
Gutierrez P, Closa D, Piner R, Bulbena O, Menendez R, Torres A (2010) Macrophage activation in exacerbated COPD with and without community-acquired pneumonia. Eur Respir J 36:285–291. https://doi.org/10.1183/09031936.00118909
Dagher R, Copenhaver AM, Besnard V, Berlin A, Hamidi F, Maret M, Wang J, Qu X, Shrestha Y, Wu J et al (2020) IL-33-ST2 axis regulates myeloid cell differentiation and activation enabling effective club cell regeneration. Nat Commun 11:4786. https://doi.org/10.1038/s41467-020-18466-w
Kondo Y, Yoshimoto T, Yasuda K, Futatsugi-Yumikura S, Morimoto M, Hayashi N, Hoshino T, Fujimoto J, Nakanishi K (2008) Administration of IL-33 induces airway hyperresponsiveness and goblet cell hyperplasia in the lungs in the absence of adaptive immune system. Int Immunol 20:791–800. https://doi.org/10.1093/intimm/dxn037
Ishinaga H, Kitano M, Toda M, D’Alessandro-Gabazza CN, Gabazza EC, Shah SA, Takeuchi K (2017) Interleukin-33 induces mucin gene expression and goblet cell hyperplasia in human nasal epithelial cells. Cytokine 90:60–65. https://doi.org/10.1016/j.cyto.2016.10.010
Holcomb IN, Kabakoff RC, Chan B, Baker TW, Gurney A, Henzel W, Nelson C, Lowman HB, Wright BD, Skelton NJ et al (2000) FIZZ1, a novel cysteine-rich secreted protein associated with pulmonary inflammation, defines a new gene family. EMBO J 19:4046–4055. https://doi.org/10.1093/emboj/19.15.4046
Chen Y, Zhang X (2017) Pivotal regulators of tissue homeostasis and cancer: macrophages. Exp Hematol Oncol 6:23. https://doi.org/10.1186/s40164-017-0083-4
Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A (2008) IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest 118:534–544. https://doi.org/10.1172/JCI33194
Sun X, Hou T, Cheung E, Iu TN, Tam VW, Chu IM, Tsang MS, Chan PK, Lam CW, Wong CK (2020) Anti-inflammatory mechanisms of the novel cytokine interleukin-38 in allergic asthma. Cell Mol Immunol 17:631–646. https://doi.org/10.1038/s41423-019-0300-7
Gavino AC, Nahmod K, Bharadwaj U, Makedonas G, Tweardy DJ (2016) STAT3 inhibition prevents lung inflammation, remodeling, and accumulation of Th2 and Th17 cells in a murine asthma model. Allergy 71:1684–1692. https://doi.org/10.1111/all.12937
Acknowledgements
We thank Yongbing Ba and Xuan Tang from OE Biotech Co., Ltd (Shanghai, China) for the assistance with bioinformatics analysis.
Funding
This study was supported by the National Natural Science Foundation of China (81773394, 82070001, 81970001), Natural Science Foundation of Tianjin (20JCQNJC01790, 18ZXDBSY00150), Science and Technology Planning Project of Tianjin Jinnan District (20200118), and the Haihe Hospital Fund of China (HHYY-202008). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization, HC; investigation, YL, QZ, LL; formal analysis, YL, QZ, LL, KL, XL, JW, QW, ZD, HJ, HC; data curation, YL, QZ, LL, DH, KL, XL, JW, QW, ZD, writing—original draft preparation, YL, HC, writing—review and editing, YL, HC; supervision, HC; project Administration, HC. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Ethics approval
This study was conducted in line with the ethical principles and was approved by the Animal Care and Use Committee of Haihe Hospital, Tianjin University (2021HHKT(A)-006). Human lung specimens were obtained from the Research Biorepository under the approval of the Research Ethics Committee of Haihe Hospital, Tianjin University (2021HHKT-018).
Consent to participate
The donors of the lung samples in this study have provided written informed consent.
Consent to publish
The donors of the lung samples in this study provided informed consent for publication of the images in Fig. 1c and d.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Y., Zhang, Q., Li, L. et al. LKB1 deficiency upregulates RELM-α to drive airway goblet cell metaplasia. Cell. Mol. Life Sci. 79, 42 (2022). https://doi.org/10.1007/s00018-021-04044-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-021-04044-w