Abstract
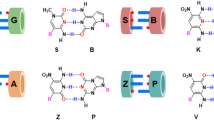
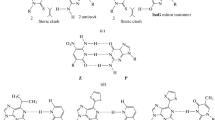
Genetic alphabet expansion is a novel concept to expand the letters of DNA, RNA, and proteins in the central dogma. This concept allows increases in the genetic information complexity and the functionalities of these biomolecules, by introducing additional components via biological systems in the programmed manner encoded on DNA with additional letters. To apply this concept to current biotechnology systems, the development of artificial extra base pairs (unnatural base pairs, UBPs) that function in replication, transcription, and/or translation is a prerequisite milestone. Several UBPs have recently been developed and applied to qPCR methods, DNA aptamer generation, site-specific labeling of RNAs, semisynthetic organism (SSO) creation, and protein synthesis involving non-standard amino acids. The introduction of hydrophobic components significantly increases the affinity of DNA aptamers to target proteins. The SSO systems with six-letter DNA/RNA produce novel functional proteins containing non-standard amino acids. Here, the development of UBPs and their applications to qPCR, DNA aptamers, and RNA modifications are discussed by focusing on UBP studies in nucleic acids.
Similar content being viewed by others
References
Benner SA, Karalkar NB, Hoshika S, Laos R, Shaw RW, Matsuura M, Fajardo D, Moussatche P (2016) Alternative Watson-Crick synthetic genetic systems. Cold Spring Harb Perspect Biol 8:a023770
Biondi E, Lane JD, Das D, Dasgupta S, Piccirilli JA, Hoshika S, Bradley KM, Krantz BA, Benner SA (2016) Laboratory evolution of artificially expanded DNA gives redesignable aptamers that target the toxic form of anthrax protective antigen. Nucleic Acids Res 44:9565–9577
Dien VT, Holcomb M, Feldman AW, Fischer EC, Dwyer TJ, Romesberg FE (2018a) Progress toward a semi-synthetic organism with an unrestricted expanded genetic alphabet. J Am Chem Soc 140:16115–16123
Dien VT, Morris SE, Karadeema RJ, Romesberg FE (2018b) Expansion of the genetic code via expansion of the genetic alphabet. Curr Opin Chem Biol 46:196–202
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Eremeeva E, Herdewijn P (2019) Non canonical genetic material. Curr Opin Biotechnol 57:25–33
Feldman AW, Romesberg FE (2018) Expansion of the genetic alphabet: a chemist’s approach to synthetic biology. Acc Chem Res 51:394–403
Flamme M, Figazzolo C, Gasser G, Hollenstein M (2021) Enzymatic construction of metal-mediated nucleic acid base pairs. Metallomics 13:mfab016
Freund N, Furst M, Holliger P (2022) New chemistries and enzymes for synthetic genetics. Curr Opin Biotechnol 74:129–136
Futami K, Kimoto M, Lim YWS, Hirao I (2019) Genetic alphabet expansion provides versatile specificities and activities of unnatural-base DNA aptamers targeting cancer cells. Mol Ther Nucleic Acids 14:158–170
Hamashima K, Soong YT, Matsunaga KI, Kimoto M, Hirao I (2019) DNA sequencing method including unnatural bases for DNA aptamer generation by genetic alphabet expansion. ACS Synth Biol 8:1401–1410
Hikida Y, Kimoto M, Yokoyama S, Hirao I (2010) Site-specific fluorescent probing of RNA molecules by unnatural base-pair transcription for local structural conformation analysis. Nat Protoc 5:1312–1323
Hirao I, Kawai G, Yoshizawa S, Nishimura Y, Ishido Y, Watanabe K, Miura K (1994) Most compact hairpin-turn structure exerted by a short DNA fragment, d(GCGAAGC) in solution: an extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res 22:576–582
Hirao I, Ohtsuki T, Kimoto M, Ishikawa M, Mitsui T, Yokoyama S (2001) Unnatural base pairs for specific transcription. Proc Natl Acad Sci U S A 98:4922–4925
Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni T, Nakayama H, Takio K, Yabuki T, Kigawa T, Kodama K, Yokogawa T, Nishikawa K, Yokoyama S (2002) An unnatural base pair for incorporating amino acid analogs into proteins. Nat Biotechnol 20:177–182
Hirao I, Harada Y, Kimoto M, Mitsui T, Fujiwara T, Yokoyama S (2004) A two-unnatural-base-pair system toward the expansion of the genetic code. J Am Chem Soc 126:13298–13305
Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S (2006) An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat Methods 3:729–735
Hirao I, Mitsui T, Kimoto M, Yokoyama S (2007) An efficient unnatural base pair for PCR amplification. J Am Chem Soc 129:15549–15555
Hoshika S, Leal NA, Kim MJ, Kim MS, Karalkar NB, Kim HJ, Bates AM, Watkins Jr NE, SantaLucia HA, Meyer AJ, DasGupta S, Piccirilli JA, Ellington AD, SantaLucia Jr J, Georgiadis MM, Benner SA (2019) Hachimoji DNA and RNA: a genetic system with eight building blocks. Science 363:884–887
Ishizuka T, Kimoto M, Sato A, Hirao I (2012) Site-specific functionalization of RNA molecules by an unnatural base pair transcription system via click chemistry. Chem Commun 48:10835–10837
Jaffrey SR (2018) RNA-based fluorescent biosensors for detecting metabolites in vitro and in living cells. Adv Pharmacol 82:187–203
Karalkar NB, Benner SA (2018) The challenge of synthetic biology. Synthetic Darwinism and the aperiodic crystal structure. Curr Opin Chem Biol 46:188–195
Kimoto M, Hirao I (2017) Unique thermal stability of unnatural hydrophobic Ds bases in double-stranded DNAs. ACS Synth Biol 6:1944–1951
Kimoto M, Hirao I (2020) Genetic alphabet expansion technology by creating unnatural base pairs. Chem Soc Rev 49:7602–7626
Kimoto M, Hirao I (2022) Genetic code engineering by natural and unnatural base pair systems for the site-specific incorporation of non-standard amino acids into proteins. Front Mol Biosci 9:851646
Kimoto M, Mitsui T, Harada Y, Sato A, Yokoyama S, Hirao I (2007) Fluorescent probing for RNA molecules by an unnatural base-pair system. Nucleic Acids Res 35:5360–5369
Kimoto M, Kawai R, Mitsui T, Yokoyama S, Hirao I (2009) An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res 37:e14
Kimoto M, Mitsui T, Yamashige R, Sato A, Yokoyama S, Hirao I (2010a) A new unnatural base pair system between fluorophore and quencher base analogues for nucleic acid-based imaging technology. J Am Chem Soc 132:15418–15426
Kimoto M, Mitsui T, Yokoyama S, Hirao I (2010b) A unique fluorescent base analogue for the expansion of the genetic alphabet. J Am Chem Soc 132:4988–4989
Kimoto M, Yamashige R, Yokoyama S, Hirao I (2012) PCR amplification and transcription for site-specific labeling of large RNA molecules by a two-unnatural-base-pair system. J Nucleic Acids 2012:230943
Kimoto M, Hikida Y, Hirao I (2013a) Site-specific functional labeling of nucleic acids by in vitro replication and transcription using unnatural base pair systems. Isr J Chem 53:450–468
Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I (2013b) Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol 31:453–457
Kimoto M, Nakamura M, Hirao I (2016) Post-ExSELEX stabilization of an unnatural-base DNA aptamer targeting VEGF165 toward pharmaceutical applications. Nucleic Acids Res 44:7487–7494
Kimoto M, Shermane Lim YW, Hirao I (2019) Molecular affinity rulers: systematic evaluation of DNA aptamers for their applicabilities in ELISA. Nucleic Acids Res 47:8362–8374
Kimoto M, Soh SHG, Hirao I (2020) Sanger gap sequencing for genetic alphabet expansion of DNA. Chembiochem 21:2287–2296
Kimoto M, Soh SHG, Tan HP, Okamoto I, Hirao I (2021) Cognate base-pair selectivity of hydrophobic unnatural bases in DNA ligation by T4 DNA ligase. Biopolymers 112:e23407
Lavergne T, Lamichhane R, Malyshev DA, Li Z, Li L, Sperling E, Williamson JR, Millar DP, Romesberg FE (2016) FRET characterization of complex conformational changes in a large 16S ribosomal RNA fragment site-specifically labeled using unnatural base pairs. ACS Chem Biol 11:1347–1353
Ledbetter MP, Karadeema RJ, Romesberg FE (2018) Reprograming the replisome of a semisynthetic organism for the expansion of the genetic alphabet. J Am Chem Soc 140:758–765
Ledbetter MP, Craig JM, Karadeema RJ, Noakes MT, Kim HC, Abell SJ, Huang JR, Anderson BA, Krishnamurthy R, Gundlach JH, Romesberg FE (2020) Nanopore sequencing of an expanded genetic alphabet reveals high-fidelity replication of a predominantly hydrophobic unnatural base pair. J Am Chem Soc 142:2110–2114
Lee KH, Kimoto M, Kawai G, Okamoto I, Fin A, Hirao I (2022) Dye-conjugated Spinach RNA by genetic alphabet expansion. Chemistry 28:e202104396
Malyshev DA, Seo YJ, Ordoukhanian P, Romesberg FE (2009) PCR with an expanded genetic alphabet. J Am Chem Soc 131:14620–14621
Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa Jr IR, Romesberg FE (2014) A semi-synthetic organism with an expanded genetic alphabet. Nature 509:385–388
Manandhar M, Chun E, Romesberg FE (2021) Genetic code expansion: inception, development, commercialization. J Am Chem Soc 143:4859–4878
Marx A, Betz K (2020) The structural basis for processing of unnatural base pairs by DNA polymerases. Chemistry 26:3446–3463
Matsunaga K, Kimoto M, Hanson C, Sanford M, Young HA, Hirao I (2015) Architecture of high-affinity unnatural-base DNA aptamers toward pharmaceutical applications. Sci Rep 5:18478
Matsunaga K, Kimoto M, Hirao I (2017) High-affinity DNA aptamer generation targeting von Willebrand factor A1-domain by genetic alphabet expansion for systematic evolution of ligands by exponential enrichment using two types of libraries composed of five different bases. J Am Chem Soc 139:324–334
Matsunaga K, Kimoto M, Lim VW, Tan HP, Wong YQ, Sun W, Vasoo S, Leo YS, Hirao I (2021a) High-affinity five/six-letter DNA aptamers with superior specificity enabling the detection of dengue NS1 protein variants beyond the serotype identification. Nucleic Acids Res 49:11407–11424
Matsunaga K, Kimoto M, Lim VW, Thein TL, Vasoo S, Leo YS, Sun W, Hirao I (2021b) Competitive ELISA for a serologic test to detect dengue serotype-specific anti-NS1 IgGs using high-affinity UB-DNA aptamers. Sci Rep 11:18000
McMinn DL, Ogawa AK, Wu Y, Liu J, Schultz PG, Romesberg FE (1999) Efforts toward expansion of the genetic alphabet: DNA polymerase recognition of a highly stable, self-pairing hydrophobic base. J Am Chem Soc 121:11585–11586
Mitsui T, Kitamura A, Kimoto M, To T, Sato A, Hirao I, Yokoyama S (2003) An unnatural hydrophobic base pair with shape complementarity between pyrrole-2-carbaldehyde and 9-methylimidazo[(4,5)-b]pyridine. J Am Chem Soc 125:5298–5307
Mitsui T, Kimoto M, Kawai R, Yokoyama S, Hirao I (2007) Characterization of fluorescent, unnatural base pairs. Tetrahedron 63:3528–3537
Morales JC, Kool ET (1998) Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat Struct Biol 5:950–954
Morales JC, Kool ET (1999) Minor groove interactions between polymerase and DNA: more essential to replication than Watson-Crick hydrogen bonds? J Am Chem Soc 121:2323–2324
Morohashi N, Kimoto M, Sato A, Kawai R, Hirao I (2012) Site-specific incorporation of functional components into RNA by an unnatural base pair transcription system. Molecules 17:2855–2876
Oh J, Shin J, Unarta IC, Wang W, Feldman AW, Karadeema RJ, Xu L, Xu J, Chong J, Krishnamurthy R, Huang X, Romesberg FE, Wang D (2021) Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II. Nat Chem Biol 17:906–914
Ohtsuki T, Kimoto M, Ishikawa M, Mitsui T, Hirao I, Yokoyama S (2001) Unnatural base pairs for specific transcription. Proc Natl Acad Sci U S A 98:4922–4925
Okamoto I, Miyatake Y, Kimoto M, Hirao I (2016) High fidelity, efficiency and functionalization of Ds-Px unnatural base pairs in PCR amplification for a genetic alphabet expansion system. ACS Synth Biol 5:1220–1230
Rappaport HP (1993) Replication of the base pair 6-thioguanine/5-methyl-2-pyrimidine with the large Klenow fragment of Escherichia coli DNA polymerase I. Biochemistry 32:3047–3057
Rich A (1962) Problems of evolution and biochemical information transfer. In: Kasha M, Pullman B (eds) Horizons in biochemistry. Academic Press, New York, pp 103–126
Sefah K, Yang Z, Bradley KM, Hoshika S, Jimenez E, Zhang L, Zhu G, Shanker S, Yu F, Turek D, Tan W, Benner SA (2014) In vitro selection with artificial expanded genetic information systems. Proc Natl Acad Sci U S A 111:1449–1454
Sherrill CB, Marshall DJ, Moser MJ, Larsen CA, Daude-Snow L, Jurczyk S, Shapiro G, Prudent JR (2004) Nucleic acid analysis using an expanded genetic alphabet to quench fluorescence. J Am Chem Soc 126:4550–4556
Someya T, Ando A, Kimoto M, Hirao I (2015) Site-specific labeling of RNA by combining genetic alphabet expansion transcription and copper-free click chemistry. Nucleic Acids Res 43:6665–6676
Switzer C, Moroney SE, Benner SA (1989) Enzymatic incorporation of a new base pair into DNA and RNA. J Am Chem Soc 111:8322–8323
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Wojciechowski F, Leumann CJ (2011) Alternative DNA base-pairs: from efforts to expand the genetic code to potential material applications. Chem Soc Rev 40:5669–5679
Yamashige R, Kimoto M, Takezawa Y, Sato A, Mitsui T, Yokoyama S, Hirao I (2012) Highly specific unnatural base pair systems as a third base pair for PCR amplification. Nucleic Acids Res 40:2793–2806
Yamashige R, Kimoto M, Okumura R, Hirao I (2018) Visual detection of amplified DNA by polymerase chain reaction using a genetic alphabet expansion system. J Am Chem Soc 140:14038–14041
Yang Z, Chen F, Alvarado JB, Benner SA (2011) Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J Am Chem Soc 133:15105–15112
Yaren O, McCarter J, Phadke N, Bradley KM, Overton B, Yang Z, Ranade S, Patil K, Bangale R, Benner SA (2021) Ultra-rapid detection of SARS-CoV-2 in public workspace environments. PLoS One 16:e0240524
Yoshizawa S, Ueda T, Ishido Y, Miura K, Watanabe K, Hirao I (1994) Nuclease resistance of an extraordinarily thermostable mini-hairpin DNA fragment, d(GCGAAGC) and its application to in vitro protein synthesis. Nucleic Acids Res 22:2217–2221
Zhang L, Yang Z, Sefah K, Bradley KM, Hoshika S, Kim MJ, Kim HJ, Zhu G, Jimenez E, Cansiz S, Teng IT, Champanhac C, McLendon C, Liu C, Zhang W, Gerloff DL, Huang Z, Tan W, Benner SA (2015) Evolution of functional six-nucleotide DNA. J Am Chem Soc 137:6734–6737
Zhang L, Yang Z, Le Trinh T, Teng IT, Wang S, Bradley KM, Hoshika S, Wu Q, Cansiz S, Rowold DJ, McLendon C, Kim MS, Wu Y, Cui C, Liu Y, Hou W, Stewart K, Wan S, Liu C, Benner SA, Tan W (2016) Aptamers against cells overexpressing glypican 3 from expanded genetic systems combined with cell engineering and laboratory evolution. Angew Chem Int Ed Engl 55:12372–12375
Zhang Y, Lamb BM, Feldman AW, Zhou AX, Lavergne T, Li L, Romesberg FE (2017a) A semisynthetic organism engineered for the stable expansion of the genetic alphabet. Proc Natl Acad Sci U S A 114:1317–1322
Zhang Y, Ptacin JL, Fischer EC, Aerni HR, Caffaro CE, San Jose K, Feldman AW, Turner CR, Romesberg FE (2017b) A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551:644–647
Zhang L, Wang S, Yang Z, Hoshika S, Xie S, Li J, Chen X, Wan S, Li L, Benner SA, Tan W (2020) An aptamer-nanotrain assembled from six-letter DNA delivers doxorubicin selectively to liver cancer cells. Angew Chem Int Ed Engl 59:663–668
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Section Editor information
Rights and permissions
Copyright information
© 2023 Springer Nature Singapore Pte Ltd.
About this entry
Cite this entry
Kimoto, M., Hirao, I. (2023). Genetic Alphabet Expansion of Nucleic Acids. In: Sugimoto, N. (eds) Handbook of Chemical Biology of Nucleic Acids. Springer, Singapore. https://doi.org/10.1007/978-981-16-1313-5_48-1
Download citation
DOI: https://doi.org/10.1007/978-981-16-1313-5_48-1
Received:
Accepted:
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-1313-5
Online ISBN: 978-981-16-1313-5
eBook Packages: Springer Reference Chemistry and Mat. ScienceReference Module Physical and Materials ScienceReference Module Chemistry, Materials and Physics