Abstract
Recent advances in single-molecule imaging have resulted in a series of discoveries regarding characteristic behavior and dynamics of individual molecules. Among the single-molecule imaging techniques, fluorescence resonance energy transfer (FRET) measurement is relatively easy to set up, yet is a powerful method; it can visualize substrate binding and dissociation as well as intramolecular structural changes within a single molecule in real time. Here, we first review single-molecule fluorescence imaging techniques that open a way to establish single-molecule FRET (smFRET) measurement. Then, we describe two examples of the characteristic dynamics of individual molecules revealed by smFRET: antibiotic-mediated protein translation inhibition and the intramolecular structural changes in CRISPR-Cas9, a versatile genome-editing tool. Finally, we introduce some of the latest advances in smFRET technique.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Fluorescence resonance energy transfer
- Molecular biophysics
- Single-molecule FRET
- Aminoglycoside antibiotics
- Gene-editing techniques
- CRISPR_Cas9
FRET techniques have been widely used for measuring the dynamics of biomolecules because of its high sensitivity as a nanoscale distance sensor. Between two closely located fluorescent molecules, energy in an excited donor fluorescent probe is resonantly transferred to an adjacent acceptor fluorescent probe, thereby decreasing the donor’s fluorescence intensity and increasing the acceptor’s fluorescence intensity. The efficiency of this energy transfer is inversely proportional to the sixth power of the distance between the two fluorescent molecules. Accordingly, FRET is an extremely sensitive measurement system for detecting changes in the distance between two fluorescent probes, particularly around what is called the Förster distance, namely, a distance that yields a FRET efficiency of 0.5 (4–7 nm for a pair of typical fluorescent probes) (Lakowicz 2006). As such, FRET measurement is ideally suited for detecting changes in the distance between domains or subunits within a protein or nucleic acids during conformational changes. Moreover, based on the ratio of fluorescence intensities of two fluorescent molecules, it can achieve high signal-to-noise ratio in measurements of binding and dissociation reactions compared with measurements involving a single fluorescent molecule. These advantages have made FRET an extensively used technique for researching the dynamics of biomolecules.
Conventional bulk FRET measurements, however, only yield mean measurement values of a large number of molecules. Therefore, these measurements are unable to extract information on the distribution of multiple molecules. The development of single-molecule imaging technologies, capable of distinguishing fluorescence intensities from individual molecules, has overcome this limitation. In combination with the single-molecule imaging techniques, FRET measurements are able to distinguish the state of each molecule in real time. This combination has led to the discoveries of diversities in dynamics and states of biomolecules in the same conditions.
In this chapter, we first outlines the single-molecule imaging techniques, which provide the basis for establishing smFRET measurement, then presents examples of protein dynamics research employing smFRET, and finally introduces some state-of-the-art smFRET applications.
1 Single-Molecule Fluorescence Imaging
To detect single molecule fluorescent probes using optical microscopy, the techniques to reduce background light combined with an intense illumination and highly sensitive camera systems are required. By the late 1980s, illumination and imaging systems had already been refined to the point of theoretically being able to detect single molecules of fluorescent probes. Nevertheless, the influence of background light from non-observed fluorescent probes had stalled the development of a system capable of identifying fluorescence from a single molecule. In 1990, Shera et al. succeeded in distinguishing fluorescent signals emitted from a single fluorescent probe molecule using pulsed light to induce photoexcitation of a low-concentration fluorescent probe solution streaming through a flow cell (Shera et al. 1990). However, this technique is arguably more pertinent to classifying as flow cytometry rather than microscopic imaging. Later, in 1993, Betzig et al. paved the way for the technology of single-molecule fluorescence imaging by employing near-field scanning optical microscopy (Betzig and Chichester 1993). Their method took advantage of the phenomenon in which illuminating a small hole with a diameter less than the wavelength of the light results in the light emerging only in the immediate vicinity of the hole. By limiting the area of illumination to a minimum, that is, reducing the background light from fluorescent probes outside the observation area, they achieved single-molecule fluorescence imaging. Thereafter, total internal reflection fluorescence microscopy (TIRFM) also has realized single-molecule fluorescence imaging by employing a similar strategy (Funatsu et al. 1995). TIRFM uses evanescent waves that occur in the immediate vicinity (~200 nm) of the interfacial surface between glass and water. Thus, only fluorescent probes present within ~200 nm from the glass surface were illuminated under TIRFM (Axelrod 1981). Compared with the near-field scanning microscopy, TIRFM is easy to set up and eligible for high-speed imaging. TIRFM requires only a standard fluorescence laser microscope with the ability to adjust the incidence angle of excitation light. Moreover, unnecessity of the scanning process makes TIRFM capable to swiftly capture images of an extensive area in a single shot. These two advantages have contributed to the widespread usage of TIRFM in the single-molecule imaging field. Today, modified epifluorescence microscopy (Sase et al. 1995), confocal microscopy (Nie et al. 1994), oblique illumination microscopy (Tokunaga et al. 2008), and light sheet microscopy (Ritter et al. 2010), which have a deeper range of photoexcitation than TIRFM, can also be applied to single-molecule imaging. Furthermore, the availability of increasingly bright commercial light-emitting diodes means that laser lighting equipment is no longer a prerequisite. These technical advances provide many options for building up a single-molecule microscopy appropriate for each experiment.
Although the above-mentioned high-speed single-molecule imaging techniques incorporate methods to reduce background light, it is necessary to limit the concentrations of fluorescent probes in solution up to the nanomolar order to distinguish single molecules from each other. Any higher concentrations render it difficult to distinguish single molecules. However, most biomolecules have a dissociation constant within the micromolar range, necessitating measurement at concentrations approximately 1000 times higher than the maximum detection limit of TIRFM techniques. To address this issue, zero-mode waveguides (ZMWs) have been developed (Levene et al. 2003). A ZMW is an optical wave guide that uses a glass foundation onto which a metallic lamina with numerous pores with a diameter of less than 100 nm is attached through vapor deposition. In a ZMW, only fluorescent probes present very close to the glass surface, approximately 10–20 nm in depth, are photoexcited. Therefore, ZMWs method can distinguish single-molecule signals under far higher fluorescent probe concentrations compared with TIRFM (Fig. 10.1). Indeed, Uemura et al. succeeded in visualizing the dynamics of tRNA that binds and dissociates from single-molecule ribosomes in the presence of fluorescently labeled tRNA at a high concentration of 2 μM during translation, thereby resolving a long-standing mystery regarding the timing of tRNA binding and dissociation from the three tRNA binding sites (Uemura et al. 2010). In addition to the capability of single-molecule imaging in the presence of high concentration probes, high-throughput data acquisition ability makes ZMWs prominent among the single-molecule imaging techniques. A large number of small pores in a ZMWs cell enables the simultaneous and parallel acquisition of single-molecule reaction data in huge quantities. Taking advantage of these high-throughput data acquisition capabilities, one of the third-generation sequencers now incorporate ZMWs. The single-molecule real-time sequencers of Pacific Biosciences, for instance, monitor and quantify hundreds of thousands of molecular reactions simultaneously, thus achieving a throughput that is one to ten million times faster than conventional Sanger sequencing (Perkel 2016). Therefore, using ZMWs, we can now acquire data of a large number of single molecules even in the presence of high concentration fluorescent probes.
Schematic diagrams of typical illumination systems using a fluorescence microscope
(a) Conventional epi-illumination. Fluorescent probes (magenta stars) that exist outside the focal plane are also photoexcited, making it difficult to eliminate background light
(b) Total internal reflection (TIR) illumination. Fluorescent probes that exist within the 100–200 nm distance from the glass surface are photoexcited. The fluorescent probes that exist outside of this range (white stars) do not interfere with observation
(c): Zero-mode waveguide illumination. An aluminum lamina with pores with a diameter of approximately 100 nm is attached to the glass surface through vapor deposition. Only the fluorescent probes that exist within the 10–20 nm distance from the glass surface are photoexcited. This trait gives this system the ability to measure single molecules under the presence of fluorescent probes at much higher concentrations compared with total internal reflection fluorescence microscopy
2 Molecular Dynamics of Proteins Measured by smFRET
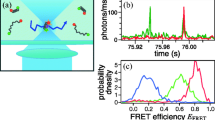
Advances in single-molecule fluorescent imaging technology have made it possible to perform smFRET measurements. smFRET measurements have revealed diversities in behavior of individual molecules in many types of bio-reactions, overturning the conventional view that molecules in same conditions show uniform reaction and dynamics. Inhibition of protein synthesis mediated by aminoglycoside antibiotics presents an example of such diversity among similar molecules revealed by smFRET. The three aminoglycoside antibiotics, apramycin, paromomycin and gentamycin, are known to inhibit protein synthesis by binding to the A site of the bacterial ribosomal small subunit. Until recently, it remained unclear which step of the ribosome-mediated protein translation process these antibiotics inhibit. Tsai et al. explored steps inhibited by these antibiotics by visualizing behavior of individual ribosomes with smFRET and ZMWs methods (Tsai et al. 2013). The major conformational changes in ribosomes (rotation of the large and small subunits) and tRNA selection processes at the A site were monitored by smFRET in ZMW cells. As a result, Tsai et al. found that apramycin inhibited the translocation of tRNA from the A site, but not affecting the major conformational change in ribosome that precedes the tRNA translocation. Meanwhile, paromomycin and gentamicin induced non-cognate tRNA binding and inhibited the ribosomal conformational change (Fig. 10.2). These results obtained from smFRET measurements clarify the detailed steps of protein synthesis, and reveal the diverse inhibition mechanisms of the antibiotics in the same type.
Action mechanism of aminoglycoside antibiotics revealed by monomolecular FRET measurement
Normally, when a second aminoacyl tRNA binds to a ribosome, the large and small ribosomal subunits rotate. This is followed by the translocation of tRNA, with the ribosome returning to the original conformation. Fluorescently labeling of the two subunits visualizes this inter-subunit rotation. smFRET measurements have revealed that paromomycin and gentamicin inhibit the first rotation and apramycin inhibits the tRNA translocation and the second ribosomal rotation, which returns the ribosome to its original conformation (Tsai et al. 2013)
smFRET measurement is an exceptionally versatile technique, capable of tracking the dynamics of not only ribosomes but also many other molecules in real time. Next, we present some of the latest findings regarding the dynamics of the RNA-guided Cas9 endonuclease, which has been recognized as a powerful genome-editing tool. Cas9 protein binds to a single-stranded guide RNA (sgRNA) to form an sgRNA–Cas9 complex, which then binds to a double-stranded DNA with a specific base sequence (NGG for S. pyogenes Cas9), known as the protospacer adjacent motif (PAM). When the DNA sequence preceding the PAM is same as that of the sgRNA, the Cas9 complex cleaves both DNA strands. Intriguingly, smFRET measurements revealed that DNA sequences scarcely affect the DNA binding rate with sgRNA-Cas9 (Singh et al. 2016). In contrast, the dissociation rate considerably increases on the introduction of mismatches proximal to the PAM sequence. This mechanism probably suppresses further reactions against off-target DNAs. Another smFRET analysis revealed heterogeneity in the behavior of the Cas9 molecules during the DNA cleavage process. In any crystal structures of the DNA-sgRNA-Cas9 ternary complex solved to date, the active site of the HNH nuclease domain in Cas9 does not attach to the DNA cleavage site (Nishimasu et al. 2014; Sternberg et al. 2015; Jiang et al. 2016). Thus, the ternary complex has been predicted to take additional temporal conformations beyond the solved crystal structures during DNA cleavage. Single-molecule measurements of intramolecular FRET between probes in the HNH domain and the domain proximal to the DNA cleavage site demonstrated that the ternary complex shows both static and fluctuating phases (Osuka et al. 2018). In the static phase, the HNH domain stays at the DNA-undocked position, which is 3-nm away from the DNA cleavage site, for an extended period. Contrarily, in the fluctuating phase, the HNH domain frequently moves between DNA-undocked, semi-docked and cleavage competent docked positions. Because the complex cleaves DNA only when the HNH is in the docked position, mutations that unstabilize the other HNH positions may enhance the nuclease activity of the Cas9. The fact that smFRET detected this temporal cleavage competent conformation, which is not solved by crystal structures, demonstrates the usefulness of smFRET as an analytical tool for investigating polymorphisms and fluctuations in protein structures (Fig. 10.3).
Dynamics of the DNA cleavage domain of Cas9. (Reproduced from Osuka et al. 2018)
smFRET measurements have revealed that HNH nuclease domain of Cas9 assumes two different phases: (a) a static phase in which the HNH domain remains stationary for an extended period (over 100 s) and (b) a fluctuating phase in which the domain moves frequently between multiple positions (Osuka et al. 2018). Only during the fluctuating phase, the Cas9 takes structure capable to cleave DNA strands. In addition, the measurement has also suggested that the HNH domain needs to temporarily translocate into the undocked position during transitions between the semi-docked and docked positions
3 Advances in smFRET Methods
The smFRET techniques presented in this chapter have been advancing further by incorporating other methodologies. For instance, attempts are being made to apply luminescent probes to observe single-molecule dynamics. Luminescent probes do not require illumination light, surmounting the obstacles of fluorescence imaging: autofluorescence, photo-damage to the samples and the strict limitations in the use of optogenetic tools. However, low brightness of the luminescent probes has limited the application of luminescent imaging. Recently, several bright and multicolor luminescent probes were developed by employing resonance energy transfer from luminescent probes to fluorescent proteins (Suzuki and Nagai 2017). The developed probes have greatly improved the sensitivity of luminescent imaging. In addition to the luminescent probes, various biosensors using FRET technique have been developed, enabling elucidation of the localization and dynamics of diverse intracellular molecules in single-molecule sensitivities.
Now, efforts are also underway to accelerate the smFRET measurement process. While conventional smFRET is able to detect the dynamics of biomolecules within 10–100 millisecond order, a recent study demonstrated that smFRET can track protein conformational changes at the microsecond level (Otosu et al. 2015). When FRET occurs, the donor fluorescent probe shows a decreased fluorescence intensity as well as a shortened cycle of time before returning to the ground state after photoexcitation (fluorescence lifetime). Compared with conventional intensity measurements, the lifetime measurements require fewer fluorescent photons to monitor FRET efficiency, and hence, can improve the temporal resolution of FRET measurements. The study using this method revealed the detailed process of conformational changes in cytochrome c protein at a sub-microsecond temporal resolution (Otosu et al. 2015).
Although the FRET techniques presented above are premised on the labeling of samples with two (or more) probes in different colors, smFRET can be measured using a fluorescent probe. Using non-fluorescent quenchers allows experimenters to perform an analysis similar to two-colored FRET measurement simply by measuring the fluorescence intensity of one fluorescent probe. When the donor fluorescent probe approaches a quencher, the fluorescence intensity decreases due to resonance energy transfer from the donor to the quencher. Therefore, one can observe conformational changes, bindings, and dissociations of labeled biomolecules simply by measuring the change in fluorescence intensity of the donor probe (Chen et al. 2012). Moreover, not only between heterologous molecules, FRET between homologous probes (referred to as homo-FRET) can be measurable. When illuminated by polarized light, only fluorescent probes in specific orientation are photoexcited and emit polarized fluorescence. If two identical fluorescent probes locate close together, because of FRET between the probes, the probes not in the photo-excitable orientation also emit fluorescence light polarized differently than that from the photoexcited donor probe, resulting in decreased anisotropy of the fluorescence. Therefore, distance changes between identical probes can be monitored by measuring the polarization components of fluorescence. This homo-FRET technique expands the application of FRET measurement in the studies of dynamics of biomolecules which can be labeled with only one-type of fluorescent probe, such as oligomerization processes of an endogenous protein (Bader et al. 2011). These advanced optical techniques have further sophisticated the smFRET method, so that we can elucidate more detailed behaviors of wider range of biomolecules.
4 Conclusion
smFRET is a particularly useful technique for investigating the behavior of individual biomolecules. Its applications have been steadily expanding by incorporating various other optical technologies. The strength of smFRET lies in its ability to track dynamic conformational changes and bindings/dissociations of ligands under near physiological conditions in real-time. Although smFRET measurements only detect distances and angles between a few positions labeled with FRET probes, this drawback can be complemented by X-ray crystal structure and electron microscopic analyses that visualize entire structures at high resolution. The appropriate combinations of technologies will further deepen our understanding of the molecular basis of biological phenomena.
References
Axelrod D (1981) Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol 89(1):141–145
Bader AN, Hoetzl S, Hofman EG, Voortman J, van Bergen en Henegouwen PM, van Meer G et al (2011) Homo-FRET imaging as a tool to quantify protein and lipid clustering. ChemPhysChem 12(3):475–483
Betzig E, Chichester RJ (1993) Single molecules observed by near-field scanning optical microscopy. Science 262(5138):1422–1425
Chen J, Tsai A, Petrov A, Puglisi JD (2012) Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics. J Am Chem Soc 134(13):5734–5737
Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T (1995) Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature 374(6522):555
Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ et al (2016) Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351(6275):867–871
Lakowicz JR (2006) Principles of fluorescence spectroscopy. Springer, New York
Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW (2003) Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299(5607):682–686
Nie S, Chiu DT, Zare RN (1994) Probing individual molecules with confocal fluorescence microscopy. Science 266(5187):1018–1021
Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N et al (2014) Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156(5):935–949
Osuka S, Isomura K, Kajimoto S, Komori T, Nishimasu H, Shima T et al (2018, in press) Real-time observation of flexible domain movements in Cas9. EMBO J 37(10):e96941
Otosu T, Ishii K, Tahara T (2015) Microsecond protein dynamics observed at the single-molecule level. Nat Commun 6:7685
Perkel J (2016) Elaine Mardis and Richard Wilson, directors of the McDonnell Genome Institute at Washington University, in front of an Illumina HiSeq 2000 sequencer. BioTechniques 60(2):56–60
Ritter JG, Veith R, Veenendaal A, Siebrasse JP, Kubitscheck U (2010) Light sheet microscopy for single molecule tracking in living tissue. PLoS One 5(7):e11639
Sase I, Miyata H, Corrie J, Craik JS, Kinosita K Jr (1995) Real time imaging of single fluorophores on moving actin with an epifluorescence microscope. Biophys J 69(2):323–328
Shera EB, Seitzinger NK, Davis LM, Keller RA, Soper SA (1990) Detection of single fluorescent molecules. Chem Phys Lett 174(6):553–557
Singh D, Sternberg SH, Fei J, Doudna JA, Ha T (2016) Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat Commun 7:12778
Sternberg SH, LaFrance B, Kaplan M, Doudna JA (2015) Conformational control of DNA target cleavage by CRISPR–Cas9. Nature 527(7576):110
Suzuki K, Nagai T (2017) Recent progress in expanding the chemiluminescent toolbox for bioimaging. Curr Opin Biotechnol 48:135–141
Tokunaga M, Imamoto N, Sakata-Sogawa K (2008) Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods 5(2):159
Tsai A, Uemura S, Johansson M, Puglisi EV, Marshall RA, Aitken CE et al (2013) The impact of aminoglycosides on the dynamics of translation elongation. Cell Rep 3(2):497–508
Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD (2010) Real-time tRNA transit on single translating ribosomes at codon resolution. Nature 464(7291):1012
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this paper
Cite this paper
Shima, T., Uemura, S. (2020). Molecular Dynamics Revealed by Single-Molecule FRET Measurement. In: Toyama, Y., Miyawaki, A., Nakamura, M., Jinzaki, M. (eds) Make Life Visible. Springer, Singapore. https://doi.org/10.1007/978-981-13-7908-6_10
Download citation
DOI: https://doi.org/10.1007/978-981-13-7908-6_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-7907-9
Online ISBN: 978-981-13-7908-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)