Abstract
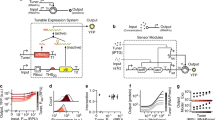
The ability to harness biomolecules as tools for systematic engineering is fundamental to future developments of biotechnology and nanotechnology. Especially suitable for such applications are nucleic acid-based circuits with predictable interactions, allowing for rational design of circuit functions and dynamics. Here, we focus on synthetic transcriptional circuits utilizing the modular architecture of nucleic acid templates and the catalytic power of natural enzymes. The programmability of dynamic behaviors for synthetic circuits is illustrated through elementary circuits such as an adapter, a bistableBistable switch, and several oscillators. Further, the effect of downstream processes on the central dynamical system illustrates the need for systematic methods of composing biomolecular circuits. We present insulating and amplifying devices as a solution for scaling up biomolecular networks much as in electronic circuits. Future applications of biomolecular programs will open up new possibilities in nanorobotics, nanomedicine, and artificial cells.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Andrianantoandro E, Basu S, Karig DK, Weiss R (2006) Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol 2:2006.0028
Atkinson MR, Savageau M, Myers J, Ninfa A (2003) Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113:597–607
Benenson Y, Shapiro E (2004) Molecular computing machines. Dekker Encyclopedia of Nanoscience and Nanotechnology. Springer, New York
Benner SA, Sismour AM (2005) Synthetic biology. Nature Rev Genet 6(7):533–543
Cheng ZF, Deutscher MP (2002) Purification and characterization of the Escherichia coli exoribonuclease RNase R: Comparison with RNase II. J Biol Chem 277:21,624–21,629
Del Vecchio D, Ninfa A, Sontag E (2008) Modular cell biology: retroactivity and insulation. Mol Syst Biol 4:161
Douglas SM, Bachelet I, Church GM (2012) A logic-gated nanorobot for targeted transport of molecular payloads. Science 335(6070):831–834
Douglas SM, Dietz H, Liedl T, Hgberg B, Graf F, Shih WM (2009) Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459:414–418
Dyson F (2007) Our biotech future. The New York Review of Books 54. http://www.nybooks.com/articles/20370. Accessed 19 July 2007
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Endy D (2005) Foundations for engineering biology. Nature 438(7067):449–453
Feng X, Hooshangi S, Chen D, Li G, Weiss R, Rabitz H (2004) Optimizing genetic circuits by global sensitivity analysis. Biophys J 87:2195–2202
Fiedler TJ, Vincent HA, Zuo Y, Gavrialov O, Malhotra A (2004) Purification and crystallization of Escherichia coli oligoribonuclease. Acta Crystallogr D Biol Crystallogr 60:736–739
Franco E, Del Vecchio D, Murray RM (2009) Design of insulating devices for in vitro synthetic circuits. In: Proceedings of the IEEE conference on decision and control, pp 4584–4589
Franco E, Friedrichs E, Kim J, Jungmann R, Murray R, Winfree E, Simmel FC (2011) Timing molecular motion and production with a synthetic transcriptional clock. Proc Natl Acad Sci USA 108(40):E784–E793
Gao Y, Wolf LK, Georgiadis RM (2006) Secondary structure effects on DNA hybridization kinetics: a solution versus surface comparison. Nucleic Acids Res 34(11):3370–3377
Goodwin BC (1965) Oscllatory behavior in enzymatic control processes. Adv Enzyme Regul 3:425–436
Grate D, Wilson C (1999) Laser-mediated, site-specific inactivation of RNA transcripts. Proc Natl Acad Sci USA 96:6131–6136
Hockenberry AJ, Jewett MC (2012) Synthetic in vitro circuits. Curr Opin Chem Biol 16:253–259
Hodgman CE, Jewett MC (2012) Cell-free synthetic biology: thinking outside the cell. Metab Eng 14(3):261–269
Jiang M, Rong M, Martin C, McAllister WT (2001) Interrupting the template strand of the T7 promoter facilitates translocation of the DNA during initiation reducing transcript slippage and the release of abortive products. J Mol Biol 310:509–522
Karkare S, Bhatnagar D (2006) Promising nucleic acid analogs and mimics: characteristic features and applications of PNA, LNA and morpholino. Appl Microbiol Biotechnol 71:575–586
Ke Y, Ong LL, Shih WM, Yin P (2012) Three-dimensional structures self-assembled from DNA bricks. Science 338(6111):1177–1183
Khalil AS, Collins JJ (2010) Synthetic biology: applications come of age. Nat Rev Genet 11(5):367–379
Kim J, Hopfield JJ, Winfree E (2004) Neural network computation by in vitro transcriptional circuits. Adv Neural Inf Proc Syst 17:681–688
Kim J, Murray R (2011) Analysis and design of a synthetic transcriptional network for exact adaptation. In: IEEE biomedical circuits and systems conference (BioCAS), pp 345–348
Kim J, White KS, Winfree E (2006) Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol Syst Biol 2:68
Kim J, Winfree E (2011) Synthetic in vitro transcriptional oscillators. Mol Syst Biol 7:465
Krishnan Y, Simmel FC (2011) Nucleic acid based molecular devices. Angew Chem Int Ed 50(14):3124–3156
Kuzyk A, Schreiber R, Fan Z, Pardatscher G, Roller EM, Hogele A, Simmel FC, Govorov AO, Liedl T (2012) DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483:311–314
Lakin MR, Parker D, Cardelli L, Kwiatkowska M, Phillips A (2012) Design and analysis of DNA strand displacement devices using probabilistic model checking. J R Soc Interface 9(72):1470–1485
Lesnik EA, Freier SM (1995) Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry 34(34):10,807–10,815
Mandell D, Kortemme T (2009) Computer-aided design of functional protein interactions. Nat Chem Biol 5(11):797–807
Markham NR, Zuker M (2005) DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res 33:577–581
Maune HT, Han SP, Barish RD, Bockrath M, III WAG, Rothemund PWK, Winfree E (2010) Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat Nanotechnol 5:61–66
Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U (2002) Network motifs: simple building blocks of complex networks. Science 298(5594):824–827
Mitarai N, Benjamin JAM, Krishna S, Semsey S, Csiszovszki Z, Masse E, Sneppen K (2009) Dynamic features of gene expression control by small regulatory RNAs. Proc Natl Acad Sci USA 106:10,655–10,659
Noireaux V, Maeda YT, Libchaber A (2011) Development of an artificial cell, from self-organization to computation and self-reproduction. Proc Natl Acad Sci USA 108:3473–3480
Phillips A, Cardelli L (2009) A programming language for composable DNA circuits. J R Soc Interface 6:S419–S436
Pinheiro AV, Han D, Shih WM, Yan H (2011) Challenges and opportunities for structural DNA nanotechnology. Nat Nanotechnol 6:763–772
Qian L, Winfree E (2011) Scaling up digital circuit computation with DNA strand displacement cascades. Science 332:1196–1201
Qian L, Winfree E, Bruck J (2011) Neural network computation with DNA strand displacement cascades. Nature 475:368–372
Rizzo J, Gifford LK, Zhang X, Gewirtz AM, Lu P (2002) Chimeric RNA–DNA molecular beacon assay for ribonuclease H activity. Mol Cell Probes 16:277–283
Rondelez Y (2012) Competition for catalytic resources alters biological network dynamics. Phys Rev Lett 108(1):018,102
Rothemund PWK (2006) Folding DNA to create nanoscale shapes and paterns. Nature 440:297–302
Rothemund PWK, Ekani-Nkodo A, Papadakis N, Kumar A, Fygenson DK, Winfree E (2004) Design and characterization of programmable DNA nanotubes. J Am Chem Soc 126(50):16,344–16, 352
Russo G, di Bernardo M, Sontag ED (2010) Global entrainment of transcriptional systems to periodic inputs. PLoS Comput Biol 6(4), e1000, 739
Saez-Rodriguez J, Kremling A, Gilles E (2005) Dissecting the puzzle of life: modularization of signal transduction networks. Comput Chem Eng 29(3):619–629
Seelig G, Soloveichik D, Zhang DY, Winfree E (2006) Enzyme-free nucleic acid logic circuits. Science 314(5805):1585–1588
Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T (2001) Cell-free translation reconstituted with purified components. Nat Biotechnol 19(8):751–755
Shin J, Noireaux V (2012) An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth Biol 1(1):29–41
Smirnakis SM, Berry MJ, Warland DK, Bialek W, Meister M (1997) Adaptation of retinal processing to image contrast and spatial scale. Nature 386:69–73
Soloveichik D, Seelig G, Winfree E (2010) DNA as a universal substrate for chemical kinetics. Proc Natl Acad Sci USA 107:5393–5398
Stoltenburg R, Reinemann C, Strehlitz B (2007) SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 24(4):381–403
Subsoontorn P, Kim J, Winfree E (2012) Ensemble bayesian analysis of bistability in a synthetic transcriptional switch. ACS Synth Biol 1:299–316
Tombelli S, Mascini M (2010) Aptamers biosensors for pharmaceutical compounds. Comb Chem High Throughput Screening 13(7):641–649
Triana-Alonso FJ, Dabrowski M, Wadzack J, Nierhaus KH (1995) Self-coded 3\(^{\prime }\)-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J Biol Chem 11:6298–6307
Tu Y, Shimizu TS, Berg HC (2008) Modeling the chemotactic response of Escherichia coli to time-varying stimuli. Proc Natl Acad Sci USA 105:14,855–14,860
Tyson JJ, Chen KC, Novak B (2003) Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol 15:221–231
Vincent HA, Deutscher MP (2006) Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem 281:29,769–29,775
Wong WW, Tsai TY, Liao JC (2007) Single-cell zeroth-order protein degradation enhances the robustness of synthetic oscillator. Mol Syst Biol 3:130
Yeung E, Kim J, Yuan Y, Gonçalves J, Murray R (2012) Quantifying crosstalk in biochemical systems. In: Proceedings of the IEEE conference on decision and control, pp 5528–5535
Yurke B, Mills AP (2003) Using DNA to power nanostructures. Genet Program Evolvable Mach 4:111–122
Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA (2011) NUPACK: analysis of nucleic acid systems. J Comput Chem 32:170–173
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Kim, J., Franco, E. (2014). Synthetic Biochemical Devices for Programmable Dynamic Behavior. In: Kulkarni, V., Stan, GB., Raman, K. (eds) A Systems Theoretic Approach to Systems and Synthetic Biology II: Analysis and Design of Cellular Systems. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-9047-5_12
Download citation
DOI: https://doi.org/10.1007/978-94-017-9047-5_12
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-9046-8
Online ISBN: 978-94-017-9047-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)