Abstract
Genes linked to major neurodegenerative diseases, including Alzheimer’s, Parkinson’s, and Huntington’s diseases, were first identified over 15 years ago, but neither a full molecular explanation for the cell loss seen in human patients nor a curative therapy has yet been achieved for any of these diseases. In most model organisms, when new hypotheses are needed to explain a cellular process, genetic screens are the tool of choice. For example, ‘synthetic lethal’ screens can lead to the identification of genes that enhance the toxicity of a particular mutation, revealing pathways critical for surviving the mutation’s effects. To date, however, genome-wide unbiased screens are not feasible in mammalian central nervous system neurons except in vitro, which fails to capture the relevant disease pathologies, and no genome-wide screens have yet been conducted in the mammalian central nervous system. We outline in this short monograph the steps needed to implement a methodology that allows for genome-wide genetic screening in the central nervous system of mice to study both normal and degenerative disease gene function.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Major Neurodegenerative Diseases
- Genome-wide Genetic Screens
- Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)
- Virus Library
- Human Gene Therapy Clinical Trials
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Genome-wide genetic screens have been used for decades in S. cerevisiae, C. elegans, and D. melanogaster to elucidate many important aspects of cell biology. Such traditional mutagenesis-based genome-wide genetic screens have been impossible to routinely perform in mice due to the prohibitively large number of mice that would be needed. However, the ability to perform such screens in the nervous system would enable the generation of new hypotheses regarding the molecular mechanisms of disease. For example, unbiased genome-wide genetic screens could reveal genes that are involved in the toxicity of disease-associated mutations, such as mutations in the huntingtin gene that are found in human Huntington’s disease patients. Such neurodegenerative disease-focused genetic screens have been attempted in S. cerevisiae, C. elegans, and D. melanogaster, but these screens by definition fail to capture the full complexity of mammalian neurons—an important point, given the widely varying susceptibility seen amongst cell types in neurodegenerative diseases. Alternatively, genome-wide genetic screens that utilize mammalian neuron-like cells have been conducted in vitro, but these screens are also unable to recapitulate the many aspects of in vivo neurons in the mammalian central nervous system (CNS). The in vivo context may be essential to many aspects of CNS biology, given for example the diversity of CNS cell types, the likely importance of both cell autonomous and non-cell autonomous factors in neurodegenerative diseases, and the known age dependency of most neurodegenerative diseases. Ideally, these screens would be done in mammalian neurons in their native cellular environment.
To bypass the difficulties associated with classical mutagenesis screening as well as the diploid nature of mammalian genomes, genome-wide short hairpin (shRNA) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) screening approaches have been applied to mammalian cells in vitro (e.g., among many others, Moffat et al. 2006; Root et al. 2006; Shalem et al. 2014; Wang et al. 2014; Zhou et al. 2014). Despite the power of these methodologies, there are many challenges to their application in vivo, especially in the CNS. Indeed, mammalian genome-wide shRNA or CRISPR genetic screens have been conducted mainly either in vitro, in transformed cell lines, or else in primary cells manipulated ex vivo and then returned in vivo (Chen et al. 2015; Graham and Root 2015). Based on the insights that have come from such studies, genome-wide genetic screening could be a powerful tool for the study of normal cellular function and degenerative disease processes in the mammalian CNS, provided that such screens are performed in the context of models that recapitulate the relevant biology. For this reason, we recently developed a genetic screening workflow that allows rapid, high-sensitivity screening in the mouse CNS for aging and neurodegenerative disease processes (Shema et al. 2015). This workflow combines the use of (1) pooled lentiviral shRNA libraries; (2) stereotaxic injection of these pools into mouse models of neurodegenerative disease and wild-type littermates; (3) incubation of injected libraries, such that shRNAs that enhance neurodegenerative disease gene toxicity lead to cell death; and (4) sequencing and analysis of the remaining shRNAs elements in all surviving cells in order to determine which constructs have enhanced cell death and thus ‘drop out’ of library representation (Fig. 1).
Genome-wide genetic screening in the mammalian CNS. Pooled viral libraries containing shRNAs, gRNAs, or cDNAs are first concentrated via ultracentrifugation to a high titer suitable for bilateral injection into the striatum (or other CNS target area) for in vivo transduction. After injection, viral payloads are allowed to integrate into the host cell genome and express for several weeks. During this time, genetic perturbations that enhance toxicity in a disease model context may enhance cell death. The targeted tissue is then carefully dissected and the genomic DNA is extracted. After PCR and sequencing of library elements, deconvolution and analysis reveals the library representation. Those genes that enhance cell death in vivo will be depleted or lost from the library (red arrows) in the mutant as compared to control animals and thus can be identified as potential modifiers of neuronal toxicity (orange barcode). These genes can then be confirmed in follow-up validation experiments
For our genetic screening workflow, we first used shRNA viral libraries, since genome-wide shRNA libraries for the mouse genome are available and have been successfully utilized in many studies. In our pilot screen, we chose to target genes that enhanced the lethality of a fragment of the mutant huntingtin gene. Huntington’s disease is the most common inherited neurodegenerative disorder, but the molecular pathways that are essential for mutant Huntingtin protein’s toxicity in vivo are not fully understood. Huntington’s disease is particularly amenable to genetic screening, as it is a monogenic disease for which several mouse models exist (Huntington’s Disease Collaborative Research Group 1993; Mangiarini et al. 1996), and the most greatly affected brain region (caudate-putamen/striatum) is a well-delineated sub-cortical structure. Since Huntington’s disease displays an aging component (Mattson and Magnus 2006), we first chose to target a set of genes that showed altered expression both in the context of normal aging and in mutant Huntingtin expression in CNS neurons. Of these genes, we identified one, Gpx6, that enhances the toxicity of mutant Huntingtin protein when its expression is reduced and that partially reverses Huntington’s disease-like symptomatology when overexpressed in mouse striatum (Shema et al. 2015). With this proof-of-principle study complete, we outline below parameters that will be essential to extend this methodology to perform genome-wide screening in the mammalian CNS.
Genome-Wide Viral Library Preparation and Delivery
Stable and long-term transduction of post-mitotic neurons by lentivirus has been in use for over 20 years (Naldini et al. 1996a, b). The available genome-wide shRNA or CRISPR guide RNA (gRNA) viral libraries described to date are typically packaged with a vesicular stomatitis virus-G (VSV-G) envelope due to resulting high stability and wide host cell range of the virus (Moffat et al. 2006; Root et al. 2006; Shalem et al. 2014; Wang et al. 2014; Zhou et al. 2014). VSV-G pseudotyping additionally enhances the neuronal tropism of lentivirus (Burns et al. 1993; Yee et al. 1994). Concentration of the initially obtained viral supernatants by ultracentrifugation yields high titers of intact VSV-G pseudotyped virus (Burns et al. 1993; Yee et al. 1994) that are essential for in vivo stereotaxic injections into the brain. As lentivirus is a relatively large virus (~100 nm), its diffusion is limited in the dense neuropil of the mammalian CNS. Given this consideration, injection parameters must be carefully optimized for each target tissue region (Cetin et al. 2006). Adeno-assisted virus (AAV) represents another potential delivery vehicle for pooled screens. As AAV is a small (~20 nm) non-enveloped virus that can be concentrated to very high titers, it is ideal for in vivo CNS delivery and, for this reason, AAV vectors have been widely used in human gene therapy clinical trials (Hocquemiller et al. 2016). Drawbacks to using AAV include its limited payload size (~4.5 kb), which limits the ability to perform cDNA overexpression screens, and the fact that the AAV serotype to be used may need to be optimized for the CNS cell type of interest.
The choice of viral library payload will depend on the experimental goals of the screening project but, in principle, cDNA, shRNA, or CRISPR gRNA libraries could all be used to interrogate CNS gene function. A recent study that compared the results of both shRNA and CRISPR/Cas9 gRNA screens to identify essential genes in a leukemia cell line found modest correlation between screen results (Morgens et al. 2016), and in some biological contexts it may be that both knockdown (shRNA or CRISPRi; Qi et al. 2013) and knockout (CRISPR) strategies should be employed to examine disease-relevant mechanisms (Deans et al. 2016).
Once a viral library is chosen and prepared, the number of cells needed for genome-wide screening should be estimated to determine the feasibility of conducting screening in the desired CNS cell population. Based on past shRNA and CRISPR gRNA screens, approximately 1000 cells should be targeted per library element, depending on the details of the screen. This number is necessary to average out noise in the assay itself, and also heterogeneity in the genetic perturbation induced in each cell, as well as inherent variability in the response of the screened cells to the perturbation. (Graham and Root 2015). Thus, for a CRISPR gRNA library that contains approximately four gRNAs per protein-coding gene, the 80,000 library elements should each be targeted to approximately 1000 cells (thus 80 million cells in total across all replicates). Reducing either biological or technical variability, for example by employing a more homogeneous cell population, can reduce the number of cells needed in each screen. The time between injection of the library and harvesting of the cells for analysis will be determined by experimental goals and could range from several days to months, depending on the rate of progression of the CNS phenotype being screened.
Interpretation of Results
As in other pooled RNAi/CRISPR screens, in CNS genome-wide screens genomic DNA is extracted from the target tissue and subjected to PCR for constant regions in the shRNA/gRNA sequences. The samples are then barcoded, pooled, sequenced, and run through deconvolution analysis to determine the representation of each individual library element. A few key factors that determine the quality and the interpretation of the results are the number of elements targeting each individual gene, if it is shRNA, gRNA or cDNA, and the depth of sequencing. A number of different methods and tools have been designed to analyze pooled screening data, and these differ based on library complexity and the type of element used to induce the perturbation. There are also a number of analytical tools that have been developed for analysis of RNAi and CRISPR genome-wide screens to assign enrichment/depletion scores, including, for example, Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK), RNAi Enrichment Gene Ranking (RIGER), and STARS, which rank shRNA or gRNA performance based on magnitude and consistency of elements for each gene that is depleted or enriched (Luo et al. 2008; Li et al. 2014; Doench et al. 2016). Another tool, Cas9 high-Throughput maximum Likelihood Estimator (casTLE), can be used to combine data of shRNA and gRNA screens to increase sensitivity (Morgens et al. 2016).
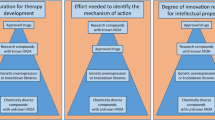
A primary genome-wide in vivo screen may yield hundreds of hits, and independent validation of these targets is necessary to confirm the assay results and the gene specificity of the observed effects and to understand the role of the genes in modifying disease phenotypes (Fig. 2). Two strategies for validation of genome-wide in vivo screening can be utilized to assess performance of the primary screen and confirm hits. Creation of sub-pool libraries allows efficient validation of several hundred potential hits. This strategy has been used to validate findings in vitro and in cells reintroduced in vivo (Chen et al. 2015). Sub-pool elements could include shRNAs or gRNAs that target genes that were unchanged in the primary screen, an additional 4–5 shRNAs or gRNAs for the primary screen hit genes, and carefully designed 9C11 controls that reveal shRNA seed-related off-target effects (Buehler et al. 2012). A second approach to validation is by interrogation of individual hits via traditional single-gene knockout/knockdown/overexpression studies. To do this, in addition to classical germline genetic perturbations, CNS viral delivery of top screen-hit validated shRNAs/gRNAs/cDNAs by stereotaxic injection can be used to rapidly introduce a single genetic perturbation, as is routinely performed in many CNS studies with AAV or retroviral vectors. This type of more traditional validation approach has the advantage that it can be used to assay various behavioral and pathological readouts of disease progression and to tease out specific biochemical pathways.
Validation of in vivo screening hits. A primary genome-wide in vivo screen is completed with at least 4–6 elements targeting a single gene, leading to libraries composed of ~80,000–120,000 elements. Validation of genes identified in the primary screen can be completed with smaller sub-pool libraries of only ~10,000–20,000 elements, which must be carefully designed to include an increased number of unique elements (~10) targeting the positive hits identified in the genome-wide screen as well as appropriate controls. These controls come in the form of elements targeting non-genomic sequences, genes unchanged in the primary screen, and C911 controls that can reveal seed-related off-target effects of hits. Sub-pool validation using a combination of multiple modalities (i.e., cDNA, gRNA and shRNA) may also be used to increase confidence in hits. Additional validation at the single-gene level can then be performed via viral transduction of two to three targeting elements and appropriate controls or else traditional knockdown/knockout/overexpression studies. Such single-gene validation is particularly important for investigation of behavioral and pathogenic readouts of disease processes as well as biochemical mechanisms underlying modification of toxicity
In addition to validation of targets from a single primary screen utilizing a particular genetic perturbation, comparison of data from two different modalities, i.e., both shRNA knockdown and gRNA knockout, or cDNA overexpression and gRNA knockout, may be beneficial. This cross-platform approach has shown to produce varying degrees of overlap in identified targets (Deans et al. 2016; Evers et al. 2016; Morgens et al. 2016), highlighting the possible utility of applying several types of perturbations in a multi-armed screen to enhance the specificity of hits or else to expand the type of hits that can be obtained (e.g., certain phenotypes may only be revealed upon gene knockdown, not knockout). While primary genome-wide cDNA screens may be challenging due to the efficiency of packaging genome-wide cDNAs into viral vectors, the potential for use in sub-pool screening of a smaller number of genes is much higher. Therefore, a combination of these techniques (cDNA overexpression, shRNA knockdown, gRNA CRISPR or CRISPRi) may yield increased sensitivity to uncover biological pathways relevant to neuronal function and dysfunction.
Future Directions
Looking forward, the ability to perform cell type-specific genome-wide genetic screens will be helpful to fully understand CNS disease mechanisms, as most neurological diseases display cell type-specific patterns of vulnerability, including the two most prevalent neurodegenerative diseases, Alzheimer’s disease and Parkinson’s disease (Mattson and Magnus 2006). The use of a conditional Cas9-expressing mouse line crossed to one that expresses Cre recombinase in the cell type of interest should allow such cell type-specific CRISPR knockout or CRISPRi gRNA screens. Conditional or inducible systems for use with mammalian retroviral vectors (Beier et al. 2011) could be useful for lentiviral-based shRNA or cDNA overexpression screens. Genome-wide genetic screening in the mammalian CNS may make it possible to interrogate molecular mechanisms linked to all the major neurodegenerative diseases and eventually to identify common vulnerability factors that may exist among these diseases, for example, aging-related and proteostasis pathways. Finally, the ability to perform genetic screening in the CNS around a non-death phenotype (e.g., biomarker expression using flow-sorting to isolate the hit cells) would greatly expand the power of genome-wide approaches.
References
Beier KT, Samson ME, Matsuda T, Cepko CL (2011) Conditional expression of the TVA receptor allows clonal analysis of descendents from Cre-expressing progenitor cells. Dev Biol 353:309–320
Buehler E, Chen YC, Martin S (2012) C911: a bench-level control for sequence specific siRNA off-target effects. PLoS One 7:e51942
Burns JC, Friedmann T, Driever W, Burrascano M, Yee JK (1993) Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA 90:8033–8037
Cetin A, Komai S, Eliava M, Seeburg PH, Osten P (2006) Stereotaxic gene delivery in the rodent brain. Nat Prot 1:3166–3173
Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, Lee H, Zhang F, Sharp PA (2015) Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160:1246–1260
Deans RM, Morgens DW, Okesli A, Pillay S, Horlbeck MA, Kampmann M, Gilbert LA, Li A, Mateo R, Smith M, Glenn JS, Carette JE, Khosla C, Bassik MC (2016) Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat Chem Biol 12:361–366
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191
Evers B, Jastrzebski K, Heijmans JP, Grernrum W, Beijersbergen RL, Bernards R (2016) CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol 34:631–633
Graham DB, Root DE (2015) Resources for the design of CRISPR gene editing experiments. Genome Biol 16:260
Hocquemiller M, Giersch L, Audrain M, Parker S, Cartier N (2016) Adeno-associated virus-based gene therapy for CNS diseases. Hum Gene Ther 27:478–496
Huntington’s Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–983
Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS (2014) MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15:554
Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn W, Lander ES, Sabatini DM, Root DE (2008) Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA 105:20380–20385
Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87:493–506
Mattson MP, Magnus T (2006) Ageing and neuronal vulnerability. Nat Rev Neurosci 7:278–294
Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283–1298
Morgens DW, Deans RM, Li A, Bassik MC (2016) Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol 34:634–636
Naldini L, Blomer U, Gage FH, Trono D, Verma IM (1996a) Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA 93:11382–11388
Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D (1996b) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183
Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM (2006) Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods 3:715–719
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87
Shema R, Kulicke R, Cowley GS, Stein R, Root DE, Heiman M (2015) Synthetic lethal screening in the mammalian central nervous system identifies Gpx6 as a modulator of Huntington’s disease. Proc Natl Acad Sci USA 112:268–272
Wang T, Wei JJ, Sabatini DM, Lander ES (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science 343:80–84
Yee JK, Miyanohara A, LaPorte P, Bouic K, Burns JC, Friedmann T (1994) A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci USA 91:9564–9568
Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W (2014) High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509:487–491
Acknowledgments
We wish to thank Dr. David E. Root and Dr. John G. Doench for useful discussions and advice. This work was supported by grants to M.H. by the JPB Foundation and NIH/NINDS.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2017 The Author(s)
About this chapter
Cite this chapter
Wertz, M.H., Heiman, M. (2017). Genome-Wide Genetic Screening in the Mammalian CNS. In: Jaenisch, R., Zhang, F., Gage, F. (eds) Genome Editing in Neurosciences. Research and Perspectives in Neurosciences. Springer, Cham. https://doi.org/10.1007/978-3-319-60192-2_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-60192-2_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-60191-5
Online ISBN: 978-3-319-60192-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)