Abstract
Ancient DNA (aDNA) promises to revolutionise our understanding of crop evolution. Wheat has been a major crop for millennia and has a particularly interesting history of domestication, dispersal, and hybridisation, summarised briefly here. We review how the fledgling field of wheat archaeogenomics has already contributed to our understanding of this complex history, revealing the diversity of wheat in ancient sites, both in terms of species and genetic composition. Congruently, ancient genomics has identified introgression events from wild relatives during wheat domestication and dispersal. We discuss the analysis of degraded aDNA in the context of large, polyploid wheat genomes and how environmental effects on preservation may limit aDNA availability in wheat. Despite these challenges, wheat archaeogenomics holds great potential for answering open questions regarding the evolution of this crop, namely its domestication, the different dispersal routes of the early domestic forms and the diversity of ancient agricultural practices. Not only will this research enhance our understanding of human history, but it will also contribute valuable knowledge about ancient selective pressures and agriculture, thus aiding in addressing present and future agricultural challenges.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
7.1 Shining a Light on the Past: The Promise of Ancient DNA
Ancient DNA (aDNA) has fostered a revolution in evolutionary genomics, as it allows direct observation of historical molecular diversity (Der Sarkissian et al. 2014). Previously, hypotheses were based solely on the observation of modern genetic diversity, which is the end effect of thousands of years of evolution, with the main caveat that the same pattern of genetic variation is often consistent with different historical scenarios (Lawson et al. 2018). The analysis of aDNA allows the genomic characterization of populations at different points in time, adding a fundamentally new dimension to evolutionary studies (Gutaker and Burbano 2017; Orlando et al. 2021).
The very first aDNA analysis was conducted on a mitochondrial sequence of a museum-preserved quagga (Higuchi et al. 1984). Since then, the field of archaeogenomics has rapidly flourished (Morozova et al. 2016), allowing for a better understanding of human, animal, and plant evolutionary history. Recent advances in this field include sedimentary, epigenetic, pathogens, and microbiome aDNA analysis (Key et al. 2020; Parducci et al. 2017; Pedersen et al. 2014; Spyrou et al. 2019; Warinner et al. 2014).
aDNA has already had a remarkable impact on our understanding of human history, shedding light on important patterns of migration (Lacan et al. 2011), admixture (Yang et al. 2020), adaptation (Marciniak and Perry 2017), population dispersal, expansion, and decline (Nielsen et al. 2017). Notably, aDNA gave fundamental contribution to our knowledge about the genetic relationships between modern humans and their extinct relatives Neanderthals (Weyrich et al. 2015) and Denisovans (Krause et al. 2010; Reich et al. 2010), the latter of which have only been identified through aDNA analysis. Similar insights have been gained in other animals, such as dogs (Botigue et al. 2017; Leathlobhair et al. 2018), cattle (Daly et al. 2018; Verdugo et al. 2019), pigs (Frantz et al. 2019), and horses (Gaunitz et al. 2018). These studies have led to a reassessment of previous evidence and an overturning of the existing narrative (Librado et al. 2021).
Now, aDNA promises a similar revolution in our understanding of how crops have been domesticated and spread around the globe, and the ways that these processes have shaped genetic diversity. By revealing how crops have adapted to new environments and what genetic diversity has been lost, aDNA can also set a basis for future breeding strategies (di Donato et al. 2018; Pont et al. 2019b). Crop archaeogenomics is still in its infancy, but aDNA from several important crops has been analysed, including maize (Ramos-Madrigal et al. 2016), barley (Mascher et al. 2016; Palmer et al. 2009), cotton (Palmer et al. 2012), bean (Trucchi et al. 2021), sunflower (Wales et al. 2018), sorghum (Smith et al. 2019), watermelon (Renner et al. 2019), and emmer wheat (Scott et al. 2019).
In this chapter, we first give a very brief overview of the history of wheat cultivation and the key genetic changes involved. The aDNA technology promises unique insights in this area. We review the wheat aDNA studies carried out so far and their contribution to understanding phenomena that have shaped wheat genomes. To conclude, we discuss the key open questions in this field and discuss the limitations posed by wheat’s large polyploid genome and idiosyncratic preservation. Our goal is to give an overview of the important answered and unanswered questions in the history of wheat cultivation and the promise of aDNA for resolving them.
7.2 A Brief History of Wheat Cultivation
Human societies have relied on wheat for thousands of years. Thus, the history of wheat domestication, geographic expansion, and cultivation has cross-disciplinary significance (Fig. 7.1). Understanding how wheat genetic diversity has been shaped also has contemporary relevance due to its continued nutritional and economic importance. Archaeogenomic studies aim to give new information about at least three key aspects of this process: domestication, dispersal, and gene flow between different wheat species. To contextualize contributions from archaeogenomics, we briefly overview these basic tenets of wheat cultivation history.
Wheat has been culturally important for millennia, and DNA extracted from ancient specimens can reveal how humans have shaped crop genetic diversity. Left: Facsimile of a vignette on the tomb of Sennedjem and Iineferti showing grain harvest in the abundant fields of the next life (painted by Charles K Wilkinson in 1922 CE, original ca. 1295–1213 BCE, public domain image from the Metropolitan Museum of Art). Right: Archaeological specimens of desiccated emmer wheat chaff from Egypt. Photo from Dorian Q. Fuller, University College London, Institute of Archaeology
7.2.1 Domestication
Wild tetraploid emmer wheat was one of the first species to be domesticated (Haas et al. 2018), during the so-called Neolithic Transition, in parallel with humans’ shift from hunting and gathering to agriculture and animal husbandry (Diamond 2002). The quintessential trait for cereal domestication is the loss of rachis brittleness: in wild cereals, the spikelets disarticulate spontaneously from the rachis upon maturity, ensuring seed dispersal and germination. In domestic cereals, the rachis is non-brittle; spikelets remain attached, allowing easier harvesting but requiring subsequent sowing in the following season in order to germinate. Because plants with a non-brittle rachis depend on human action for dispersal, this phenotype has been used to define domestication in cereals (Abbo et al. 2014; Snir et al. 2015). Loss-of-function mutations in the TtBtr1-A and TtBtr1-B genes on chromosomes 3A and 3B are the main determinants of such phenotype (Avni et al. 2017; Nave et al. 2019). Therefore, alleles at these two loci essentially distinguish wild from domesticated emmer wheat. Other traits that are favourable in the human-mediated environment and most likely deleterious in a wild environment (Kantar et al. 2017; Purugganan and Fuller 2009) give a more broad definition of the “domestication syndrome” (Larson et al. 2014), like the loss of seed dormancy and larger seed sizes (Haas et al. 2018; Zohary 2013).
Wild emmer wheat has a very restricted distribution, growing only in the Fertile Crescent region of Southwest (SW) Asia (Vavilov et al. 1992). The exact location of the emergence of domestic emmer has been a long-standing controversy. In the 2000s, early genetic studies started addressing this issue, with the so-called cradle of agriculture theory (Lev-Yadun et al. 2000). Further genetic studies had pointed to the Northern Fertile Crescent and specifically to the Karaca Dağ Mountain region as the centre of domestication of emmer wheat (Luo et al. 2007; Ozkan et al. 2002, 2005), mostly based on the higher similarities between the genomes of the modern domestic landraces and the wild emmer from the Northern Levant, compared to that of the Southern Levant (Avni et al. 2017).
However, this monophyletic origin has been challenged with increasing evidence that different wild populations have contributed to domestic wheats. Several authors argue that domestic emmer wheat arose from an admixed wild population and that mutations for domestication traits appeared in different chromosomes at different times and possibly in different places (Civáň et al. 2013; Jorgensen et al. 2017; Oliveira et al. 2020). This is in line with the observation that the domestic phenotype, which requires at least two independent recessive mutations, took millennia to be established (Avni et al. 2017; Fuller et al. 2014). As testified by the archaeological record, wild emmer wheat was first exploited in the Southern Levant, where increasing, even though small, proportions of phenotypically domestic emmer wheat are found at different archaeological sites as early as during Early Pre-Pottery Neolithic B (8700–8200 BCE) (Arranz-Otaegui et al. 2018). However, domesticated emmer is found in very high proportions in the Northern Levant starting from the Middle/Late Pre-Pottery Neolithic B (8200–6300 BCE) (Arranz-Otaegui et al. 2016). This indicates that wild emmer was managed (a phenomenon often regarded as “pre-domestication cultivation”) (Fuller et al. 2010) long before the domestic forms emerged, and that probably wild populations from across the Fertile Crescent contributed to the domestic pool (Feldman and Kislev 2007). The role of introgression from wild to domestic wheat has been demonstrated by several studies, e.g. (Cheng et al. 2019; Pont et al. 2019b; Przewieslik-Allen et al. 2021), even though the context in which these introgression events took place remains unknown.
Overall, archaeology and genetics point to a slow and geographically widespread domestication process in which both the Northern Levant and the Southern Levant played an important role.
7.2.2 Evolution
Domestic emmer wheat (Triticum turgidum subsp. dicoccon) gave rise to today’s most economically important wheats: tetraploid durum wheat (T. turgidum subsp. durum) and hexaploid bread wheat (T. turgidum subsp. aestivum). These descendants differ from their ancestor in one character of great agricultural importance: the free-threshing phenotype. Emmer is a hulled, non-free-threshing wheat, and the extraction of seeds from husks requires substantial mechanical processing. On the other hand, durum and bread wheat are naked and free-threshing: as the spikelets disarticulate from the rachis they fall apart, releasing the seeds without further processing. While durum wheat is tetraploid (BBAA), bread wheat is hexaploid (BBAADD) and evolved from the hybridization of tetraploid wheat with the diploid wild goatgrass (Aegilops tauschii), donor of the D subgenome (Haas et al. 2018; Pont et al. 2019a). The tetraploid that contributed the B and A subgenomes to bread wheat has been a matter of debate (Sharma et al. 2019), but considering the need for multiple mutations to determine the free-threshing phenotype, the most supported (and most parsimonious) models indicate that hybridization with A. tauschii occurred with a free-threshing tetraploid (Zhou et al. 2020).
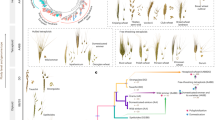
The emergence of modern wheat is therefore the result of three processes: (I) domestication of wild emmer wheat, associated with the loss of rachis brittleness; (II) crop evolution (often also referred to as crop improvement under cultivation), which includes the emergence of the free-threshing phenotype and adaptation to new ecological niches; (III) allopolyploidization between a free-threshing tetraploid with A. tauschii, giving rise to bread wheat. We summarize these changes in Fig. 7.2.
Schematic representation of the domestication and evolution of the most economically important wheats today, showing important phenotypes and the mutations that determine them. Basic information about the appearance of the different wheats in the archaeological record is given on the right. The small white hand represents the investment of human labour in processing the harvest. *For simplicity, we use the common name “durum wheat” for all free-threshing tetraploids, but other common names are used for free-threshing tetraploids, and it is not known which was involved in this allopolyploid event. This scheme is an adaptation of the model proposed by Sharma et al. (2019)
Perhaps surprisingly, hulled wheats continued to be used for thousands of years after the appearance of free-threshing durum wheat and bread wheat. The slow and regionally specific shifts in wheat usage probably reflect cultural practices and preferences (Nesbitt and Samuel 1996). Also, increasing archaeological evidence shows that early farmers relied on a wide range of other domestic wheats for their subsistence, including einkorn, spelt, and Triticum timopheevii alongside emmer and free-threshing wheats (Özbaşaran et al. 2018). This is in accordance with the evidence for intra and interspecific introgression that has been detected in modern wheat (Cheng et al. 2019; Zhou et al. 2020).
7.3 Archaeogenomics of Wheat
Wheat archaeogenomics is a powerful tool to investigate how wild wheat evolved into domestic forms and how these domestic wheat varieties adapted to different ecological niches and cultural preferences through history.
However, the limitations and the characteristics of ancient genomes have to some extent impacted the approach taken in this research field. Before high-quality reference genomes were available, most studies avoided whole-genome analysis and used a target and amplification strategy. This mitigates the challenges of a large genome but gives much less rich genomic information. Furthermore, the primers used for amplification mask the characteristic patterns of degradation that are useful for ruling out contamination by confirming the antiquity of the DNA. Unlike these amplification methods, whole-genome libraries can also be re-analysed to get more data without further destructive sampling of rare material. For these reasons, amplification approaches are no longer recommended for ancient samples (Gutaker and Burbano 2017; Prüfer and Meyer 2015).
We first overview wheat aDNA studies that use amplification and then describe the first two whole-genome analyses. Even though wheat archaeogenomics is in a germinal stage, the results have shifted our understanding of wheat genetics in important ways.
7.3.1 Target Gene Amplification
The most common use of target gene amplification has been to interrogate key genes or to identify wheat remains at the species level. The x and y copies of the Glu1 loci were often the focus of early studies. These genes, present in all wheat subgenomes, are located in the long arms of chromosome 1 and encode for the high molecular weight glutenin subunits (HMW-GSs), storage proteins present in the starchy endosperm cells of wheat. Allelic varieties in these genes impact the properties of dough for bread making. Because of its effect over bread quality, the evolution of the HMW genes can provide insights into the nature of human selective pressures during wheat evolution (Allaby et al. 1999). In this manuscript, authors surveyed these loci in a collection of modern and ancient wheats, constructed a phylogenetic tree, and obtained time estimates by using a substitution rate to calibrate the observed variation. By comparing the genetic variability for x and y copies in each genome, they were able to determine that the genetic variability in these loci for the cultivated species predates domestication, pointing to either incomplete lineage sorting, multiple domestication events, or introgression after domestication. Another study used a similar approach with the same loci to inquire about the origins of spelt (Blatter et al. 2002). They surveyed a collection of modern and ancient bread wheat and spelt specimens and determined that the high genetic variability of spelt compared to that of bread wheat in the A and B genomes are compatible with the origin of spelt being a hybridization event between bread wheat and hulled tetraploid emmer.
HMW genes have also been used to identify wheat remains at the subspecies level and inform about its dispersal. Without associated chaff, it is difficult to distinguish between free-threshing wheats (e.g. bread wheat or durum wheat). Bilgic et al. (2016) targeted the HMW promoter region in 8400-year-old specimens from a notorious Neolithic site in central Turkey, Çatalhöyük, to determine whether the genetic variability characteristic of the D genome could be recovered, as a proof of that wheat being hexaploid. The finding of HMW subunits from the A, B, and D genomes is quite remarkable, since it evidences the presence of hexaploid wheat at a very early point in time and highlights the importance of this settlement in the expansion of hexaploid wheat cultivation. Another study used the Internal Transcribed Spacer regions (ITS1 and ITS2) and the Inter-Genic Spacer region (IGS) from the nuclear ribosomal DNA for species level identification (Li et al. 2011). They also found early evidence for hexaploid wheat in Northwest China around 1760–1540 BCE.
These results highlight the high diversity of wheats consumed by humans during early agricultural expansion. Free-threshing naked wheats first appear in the archaeological record between 7000 and 5500 BCE (Feldman and Kislev 2007). Early naked wheats co-existed with domestic and wild emmer populations (Bilgic et al. 2016), giving opportunities for genetic exchange. Along with the protracted period of emmer domestication, this probably explains the higher genetic diversity on A and B subgenomes of modern bread wheat compared to the D subgenome (Cheng et al. 2019). This demonstrates how the details of agricultural history directly impact modern wheat diversity and breeding. Moreover, other wild Triticum species gave rise to domestic forms during the Neolithic. These include the diploid einkorn wheat, Triticum monococcum subsp. monococcum, that emerged from wild einkorn, T. monococcum subsp. Aegilopoides (Nesbitt and Samuel 1996), spelt (Triticum spelta), an hulled hexaploid, and tetraploid T. timopheevii (domesticated from T. timopheevii araraticum) (Wagenaar 1966), only recently classified thanks to aDNA analysis.
The position of T. timopheevii within the domestication process of wheat in SW Asia exemplifies the value of aDNA to gain insights on certain domestication processes. Briefly, due to the technical difficulties in the identification of T. timopheevii, for a long time its existence was questioned, and it was often unclassified, or ascribed to other wheat species, such as “New Glume Wheat”. Recently, archaeological remains described as “New Glume Wheat” have been designated as domestic T. timopheevii based on aDNA evidence (Czajkowska et al. 2020). The authors used the Ppd1 locus to identify G genome alleles in “New Glume Wheat” remains. This study has sparked the interest of the archaeobotanical community. Decades have passed since the first classification of an archaeological specimen to “New Glume Wheat”. It was not until numerous remains of this type of wheat were found in several Neolithic and Bronze Age archaeological sites in northern Greece and compared with other locations (Jones et al. 2000) that archaeologists were able to describe the distinctive features of this wheat (Ulaş and Fiorentino 2021). Nevertheless, identification based on grain morphology is still problematic. The identification of New Glume Wheat as domestic T. timopheevii thanks to ancient DNA analysis has had important ramifications on our understanding of the complexity of the domestication process in SW Asia and the confirmation that multiple species evolved into domestic forms, moving away from the “founder crops” theory. T. timopheevii was actually cultivated for a very long period of time in certain regions. New efforts are now being undertaken to revisit archaeobotanical assemblages and reassess the relative abundance of plant species, with the expectation that many grains classified as emmer wheat will now be classified as T. timopheevii.
The HMW loci were also used, together with the ribulose 1,5 biphosphate carboxylase (rbcL) and the chloroplast microsatellite WCT12 in the chloroplast genome to study the viability of DNA extraction on ancient plant specimens (Fernández et al. 2013). In this study, 126 grains of naked wheat in different preservation conditions (charred, partially charred, and waterlogged) were analysed (Fig. 7.3 shows different preservation conditions of ancient wheat samples). Results showed that DNA extraction from totally charred remains is virtually impossible, while DNA amplification of modern contaminants is pervasive. Unfortunately, almost all of the most ancient archaeological wheat specimens are charred, which is a severe limitation for future aDNA studies.
Examples of different preservation conditions of archaeobotanical wheat. Left: charred emmer wheat seeds from the Vinča culture in Serbia (middle/late Neolithic; c. 5400–4600/4500 BC), published in Filipovic (2014). Right: Waterlogged chaff remains of Triticum cf. durum/turgidum from the end of the 5th millennium BC at the site of Les Bagnoles. Photo by Raül Soteras, AgriChange Project, reproduced with permission
As mentioned above, one important limitation of amplification-based studies is the confidence with which one can rule out contamination. Commonly used indicators such as the fragment length distribution or deamination patterns are difficult to assess in target-specific PCR amplification studies. In addition, Allaby et al. (1999) reported PCR jumping, probably related with the shortness of some fragments. Their results showed patterns of linked diversity that did not exist in the modern pool and had to manually rearrange the observed diversity so it would match known modern haplotypes with the subsequent potential biases.
Different strategies have been used to increase confidence in the antiquity of the data. Allaby et al. replicated the results in situ with the same specimen and produced blanks with each extraction run. Czajkowska et al. (2020) performed the extractions in laboratory facilities where no wheat had been processed before, hoping to preclude contamination. Bilgic et al. (2016) processed all samples in two different facilities, so that replication of the results acts as a proof of authenticity. In spite of this, even if contamination can be ruled out, it is not possible to distinguish deamination patterns from true polymorphisms. Therefore, phylogenetic analyses and interpretation of the accumulation of variation through time should be taken with caution unless transitions (C/T or G/A SNPs) are excluded.
7.3.2 Whole-Genome Analyses
As with modern wheat samples, the genomic scale of archaeological wheat genetics has been expanded since the publication of reference genomes (Table 7.1). Nevertheless, only two studies have so far reported whole-genome sequence from archaeological wheat specimens. One has been the analysis of several bread wheat remains from China to infer dispersal into the region (Wu et al. 2019). The earliest bread wheat remains found in China date to approximately 4500 years ago in the north-western part of the country, but the most interesting aspect of its dispersal is that upon its arrival, wheat had to be adapted to a wide variety of climatic conditions. Ancient wheat from two archaeological sites within the Xinjiang winter-spring wheat zone was analysed. Even though coverage was extremely low (0.25–0.01x), the authors were able to call more than 7000 SNP sites, compare them with modern data from neighbouring regions, and provide new evidence on wheat dispersal in China, a still controversial topic. Their results were consistent with one of the routes that had been previously suggested: an early dispersal into the Qinjianh Tibetan plateau, based on the highest genetic similarities between the ancient samples and the modern ones from that region. Conversely, another ancient route that advocated for an introduction towards the eastern region was not supported. However, more data is needed to determine whether different gene pools were introduced to China and to confirm that modern landraces correspond with ancient ones from the same area.
Another whole-genome analysis of archaeobotanical specimens looked at two desiccated samples of 3000-year-old emmer wheat chaff (Fig. 7.4) from Egypt (Scott et al. 2019) to investigate early wheat dispersal and introgression from wild populations. The ancient samples were used to genotype exonic SNPs that segregate in modern accessions, at which coverage was 0.48 X after quality control, yielding approximately 100,000 high confidence genotypes. The authors used a haplotype-based approach to overcome as much as possible the limitations of aDNA analysis of polyploid species. Nearby sites that are not broken apart by recombination form co-inherited blocks called haplotypes. A “haplotype reference panel” combines information from multiple modern genomes to characterise the haplotypic variation at each genomic location (McCarthy et al. 2016). In the analysis of ancient data, when a sufficient number of genotypes can be identified within a region, it is possible to assign a known haplotype (or no known haplotype, as may be the case when ancient diversity has been lost in existing populations) to the ancient sample. At this point, non-sequenced genotypes within the region can be deduced based on haplotype assignment, a method called imputation. Haplotypes are relatively long in wheat (Walkowiak et al. 2020) because selfing tends not to break apart haplotypes as much as outcrossing. As a consequence, low coverage data is more likely to yield enough sites to assign an individual to a haplotype. This method allowed Scott et al. (2019) to identify genomic tracts tens of megabases long containing hundreds of genotypes that matched a modern sample in the haplotype reference panel. These included regions where important domestication QTLs had been identified, such that the domestication allele can be imputed and the phenotype inferred. In contrast, other genomic regions did not match anything in the haplotype reference panel.
Desiccated emmer wheat chaff from Hememiah North Spur (Egypt) 14C dated 1300–1000 BC, analysed by Scott et al. 2019. Photo by Chris J. Stevens, reproduced with permission
The data essentially confirmed that genetic changes associated with domestication were completed by 3000 years ago, prior to emmer wheat dispersal to Egypt. Nevertheless, the ancient Egyptian sample carried more “unique” haplotypes than any other domesticated sample in the dataset, indicating regions where genetic diversity has been lost. It is not yet possible to state whether this lost variation is associated with adaptation to local environmental conditions or confers other useful traits. Nevertheless, these results highlight geographic and genomic regions that may harbour genetic diversity that has been used in the past and therefore might be useful in the present and future. Moreover, while the highly repetitive nature of the wheat genome increases the chances of misalignment issues and subsequent inflated heterozygosity, Scott et al. (2019) found that the estimated heterozygosity of the ancient sample fell within the range of the modern samples. This suggests that reliable genotypes can be obtained from ancient wheat, providing appropriate quality filters are used to restrict attention to sites that do not suffer from alignment problems.
Important results from this study concern early emmer wheat dispersal. Ancient routes of dispersal generally define modern population structure and overall genetic similarity but, with the changing usage of different wheat species and the adoption of modern elite varieties, we have little grasp of historical population dispersal and replacement. Contemporary emmer wheat subpopulations (landraces) reflect the dispersal outside of SW Asia to the West (Mediterranean), to the Balkans (Eastern Europe), to Transcaucasia (Caucasus) and towards India and the Arabian peninsula (Indian Ocean) (Avni et al. 2017). The authors found that the ancient sample from Egypt resembles modern cultivars from the Indian Ocean subgroup, indicating a connection between early emmer dispersal to the East (across the Iranian Plateau and into the Indus valley) and to the South-West (Nile Valley). This is particularly interesting in light of the fact that Ethiopia currently represents a region of genetic isolation and differentiation for tetraploid wheat. This ancient Egyptian sample also has signatures of gene flow with wild populations in the Southern Levant, which could have occurred during dispersal towards Egypt or during Egyptian conquests in the Ramesside era. We expect further aDNA studies to connect historical events with changes to wheat genetics. Answering these questions will not only bring a deeper understanding of wheat evolution, but also human history, which has been intimately linked to wheat cultivation for millennia.
Overall, the field of wheat archaeogenomics has yet to reach its full potential. However, the field is primed for new advances with the availability of reference genomes and a wealth of resequenced modern landraces for comparison. While the prospects for studying DNA from charred remains are poor, many desiccated or waterlogged samples have great potential for further study. Archaeological research on waterlogged sites is increasing, which promises new material to complement the specimens currently in museums and collections.
7.4 Analysing Degraded DNA from Ancient Polyploid Wheat
Degradation and contamination are key complications for the reliable analysis of ancient DNA. To mitigate these problems, specific methods have been developed for sample preparation and downstream analysis (reviewed in Orlando et al. 2021). Even with appropriate methodology, DNA from ancient and historical samples cannot be used for all the applications that modern sequence data allows. We briefly overview these general principles of ancient DNA analysis, before discussing the specific issues posed by wheat, as all these factors should be considered during study design and analysis. We expect future methodological improvements to address these challenges, raising the possibility of resolving further important questions in the history of wheat domestication and evolution.
7.4.1 aDNA Damage
A prominent difference between ancient and modern DNA is that ancient DNA is much more fragmented prior to extraction (Fig. 7.5a). Most DNA fragmentation occurs rapidly after death (Kistler et al. 2017), as the DNA “backbone” breaks down through a process called “hydrolytic depurination”, which is biochemically predicted to occur more rapidly with exposure to water and high temperatures (Lindahl 1993). Thus, local preservation and environmental conditions are key in determining DNA yield and quality in different samples. Nevertheless, fruitful DNA sequencing has been conducted from plant tissue that is thousands of years old and from tropical and warm environments (Fornaciari et al. 2018; Mascher et al. 2016; Ramos-Madrigal et al. 2016; Renner et al. 2019). Overall, excellent DNA preservation has been reported from plant remains in desiccated and waterlogged conditions (Kistler et al. 2020).
Characteristic patterns of DNA degradation in sequence from a 3000-year-old emmer wheat sample (Scott et al. 2019). a Shows the raw distribution of fragments sizes and b shows misincorporations relative to the reference genome after alignment. In this case, the sequenced library was partially UDG treated such that the misincorporations caused by post-mortem damage are confined to a few base pairs at the fragment ends, which are removed for further analysis
Besides fragmentation, the DNA sequence itself undergoes modifications. Notably, a proportion of cytosine residues lose an amine group, becoming uracil residues, which code as thymine during sequencing (Briggs et al. 2007). This hydrolytic deamination occurs more commonly on the single stranded overhangs of the fragmented DNA molecules. As a result, when aligned to a reference genome, sequenced ancient DNA has a higher proportion of C-to-T misincorporations at the 5′ end of each fragment. Double-stranded DNA libraries will also show a higher proportion of the complementary misincorporation, G-to-A, at the 3′ end of each fragment after alignment.
These characteristic patterns of degradation found in ancient samples can be useful to the analysis, as they are proof of the sample antiquity. Therefore, the most common approach is to carry out a protocol developed for partial UDG treatment (Rohland et al. 2015). With this method, uracil-DNA-glycosylase (UDG) is used to remove uracils (Briggs et al. 2010) in the inner region of the fragments, but not at their ends. In this way, some amount of damage is maintained, but it is confined to the fragment ends (Fig. 7.5b). Similarly, the distribution of fragment lengths is used to confirm that the sequenced DNA is ancient, where large fragments may indicate contamination. Finally, paired-end sequencing of short fragments will often result in the same base pair being sequenced twice, which can be used to improve confidence in the sequence (Jonsson et al. 2014).
Standard bioinformatic protocols have been established for processing fragmented and damaged DNA. In general, standard approaches have been established for mapping short-read data to reference genomes and automated tools/pipelines are available for ancient genotypes calling for downstream analyses (Peltzer et al. 2016; Schubert et al. 2014). Common methods involve trimming off all the base pairs at the end of fragments that are potentially affected by damage (Jonsson et al. 2014) and verifying that analyses are unaffected when transitions (SNPs where the two alleles are either C/T or G/A and that can include post-mortem damage) are excluded (Korneliussen et al. 2014). We further note that “reference bias” (preferential alignment of reads carrying the same allele as the reference) is stronger in ancient data due to the shorter fragment size, so correction methods should be used (Günther and Nettelblad 2019).
For all these reasons, whole-genome sequencing has become the standard in ancient DNA studies, while PCR-based approaches are no longer considered unless for very specific goals such as genome identification, since they do not allow to verify the presence of these important patterns of post-mortem damage and to exclude contamination.
Contamination is a significant concern in ancient DNA studies. Because the amount of DNA preserved in ancient samples tends to be low, relatively small amounts of contamination from contemporary material can overwhelm the target DNA in the library (Renaud et al. 2019). Extraction and manipulation of ancient DNA therefore requires specialized facilities with protocols that minimize contamination by modern DNA (Fulton 2012). Standard practice is to create a control sequencing library without using the sample tissue (an “extraction blank”). The data from controls is analysed alongside the main sample to quantify the contamination and spurious signals likely to have been introduced during DNA extraction. Contamination can also come from microbial decomposers that invade tissues after death. A simple estimate for overall contamination is the percentage of reads that can be aligned to the reference genome of the targeted species, although other methods are available (Peyrégne and Prüfer 2020). So far, the percentage of endogenous DNA (the DNA of interest) reported in whole-genome studies of ancient plants has been high, compared to animal studies. For example, reported endogenous fractions have been 33–66% in emmer wheat (Scott et al. 2019), 5–90% in bread wheat (Wu et al. 2019), 7–54% (mean 44%) in common bean (Trucchi et al. 2021), and 70% in maize (Ramos-Madrigal et al. 2016).
Degradation and contamination limit the applications of ancient DNA, relative to modern DNA. Firstly, the fraction of endogenous DNA in well-preserved ancient DNA libraries is far below that of modern DNA (which usually is > 99%). Because endogenous fragments are short, the sequencer will often read through the DNA fragment and continue onto the adapter sequences used for library preparation. Sequenced adapter fragments must thus be discarded. Furthermore, if the sequencing has been performed for paired-ends, the forward and reverse reads will overlap (and are then collapsed into a consensus sequence). Given the low endogenous content and the short fragments, more sequence data is needed to reach reasonable coverage. Nevertheless, when small amounts of DNA are present in the sample, it may not be possible to keep sequencing to increase the coverage, since the library gradually yields diminishing returns as more duplicate reads are sequenced (Link et al. 2017). For all these reasons, coverage tends to be significantly lower in aDNA studies, when compared to the expectations for modern data.
Overall, due to low coverage and short fragments in ancient DNA, a typical approach is to identify variable sites (e.g. SNPs) using modern samples only, then use ancient DNA alignments to genotype the ancient samples. Fortunately, this approach often yields sufficient high-quality genotypes to perform analyses of interest, such as estimating genome-wide relatedness, introgression, and population genetic parameters.
7.4.2 Large Polyploid Wheat Genomes
The large genome of wheat (17 gigabases for bread wheat) implies that whole-genome sequencing of each wheat sample requires more resources compared to other organisms with smaller genomes. This cost is exacerbated in ancient DNA studies by the lower fraction of endogenous DNA, which requires further sequencing effort to obtain the same genomic coverage. In wheat, pre-designed probes are available for exons and promoters (Gardiner et al. 2019; Jordan et al. 2015), which reduce sequencing costs by enriching for sequences that are captured by the probes used. In ancient DNA, capture can enrich endogenous DNA (Hofreiter et al. 2015) but increase clonality and introduce biases towards the sequence on the probes (Ávila-Arcos et al. 2011). Exome-wide capture has not been reported for an ancient wheat. However, targeted capture might be useful to avoid repetitive regions since short aDNA fragments give little information about this class of DNA.
Ploidy and the high identity between subgenomes, estimated to be as high as 97–98%, supposes another challenge for ancient DNA studies. Even with modern samples, wheat resequencing studies can only reliably observe genomic regions that can be unambiguously aligned using the read lengths available. The shorter fragment length of ancient DNA places a practical limit on the portion of the genome that can be directly observed by mapping to reference genomes.
Heterozygosity is commonly used as an indicator of misalignment problems. Because wheat is predominantly selfing (Golenberg 1988), most sites should be homozygous in most individuals. However, various structural variants can cause reads from different genomic regions in the sample to be aligned to the same position in the reference genome (Fig. 7.6) with high mapping-quality scores, thus passing quality filters. As a consequence, sample heterozygosity will be inflated after calling genotypes. A common solution is to remove variants that are heterozygous in multiple samples, e.g. (Gardiner et al. 2019; He et al. 2019). Recent data indicates that undetected gene duplicates are common within wheat subgenomes on reference assemblies (Alonge et al. 2020). In general, polyploid wheat resequencing data will suffer from additional misalignments due to homeologous sequences on different subgenomes, but reliable genotypes can be obtained from both modern and ancient wheat provided appropriate quality filters are used to restrict attention to sites that do not suffer from alignment problems. Nevertheless, we emphasize that care should be taken when measuring heterozygosity in polyploid wheats, especially from ancient genomes. The limitations in estimating heterozygosity are unfortunate because it is heterozygosity that is a common indicator of outcrossing and genetic variation in the population, changes to which are key questions in the history of cultivation practices (Smith et al. 2019; Trucchi et al. 2021).
False heterozygosity introduced by mis-mappings to the reference. Here, we consider two genomic regions (blue and yellow), which are homeologues or duplicated regions that are relatively similar to one another. A site in each region is genotyped (coloured purple and green). In a, the sample is similar to the reference so that reads can be aligned to the correct region, and the genotype calls are all homozygous, as expected for most sites in a largely selfing species. In b, there is a difference between the reference genome and sequenced genome (indicated in grey). The sample reads from the blue genomic region in b are best aligned to the yellow region of the reference. This results in a heterozygous genotype call, while all the true genotypes are homozygous. Thus, inaccurate reference genome assemblies, deletions, insertions, or duplications can all result in spurious heterozygous genotypes
7.5 The Future of the Past: Open Questions and Prospects for Wheat aDNA
Crop archaeogenomics has already proved to be a powerful tool to investigate phenomena such as domestication, crop dispersal, and subsequent adaptation (Kistler et al. 2020; Orlando et al. 2021). Studies on bean (Trucchi et al. 2021), sunflower (Wales et al. 2019), and sorghum (Smith et al. 2019) showed that the “domestication bottleneck” (i.e. the initial loss of genetic diversity associated with domestication) may not be as intense as previously assumed. Ancient DNA analysis has been used to trace the origin of some important winemaking grape cultivars (Ramos-Madrigal et al. 2019) and brought insights on the genetic basis of potato adaptation to the European climate (Gutaker et al. 2019). In maize, adaptation to climatic constraints (selected from ancient standing variation within the domestic forms) has been identified as the main driver of modern differentiation between populations (Da Fonseca et al. 2015; Swarts et al. 2017).
7.5.1 Open Questions in Domestication
In recent years, some paradigms of domestication have been challenged by new scientific discoveries, and wheat represents a good example of such changing perspectives. Because now we know that domestic forms took thousands of years to dominate archaeological assemblages and that different wild populations seem to contribute to modern diversity, it is likely that wheat domestication was not as severe, abrupt, or geographically restricted as expected under the assumption of a “domestication bottleneck” (see Sect. 7.2). The presence of peculiar haplotypes in an ancient emmer wheat sample from Egypt showed that possibly genetic diversity has been lost after emmer wheat domestication and dispersal to Egypt (Scott et al. 2019), in line with what has been found for other species, e.g. (Trucchi et al. 2021). In the case of wheat, more ancient samples are needed to determine the association (or lack of thereof) between domestication and losses of genetic diversity.
Second, it is unclear whether there is a monophyletic “centre of domestication” for emmer wheat in the Northern Levant. The contribution of the Southern Levant gene pool to domestic emmer has been detected in several studies, but its origin remains unsolved. Whether emmer was domesticated from a proto-domestic admixed population, or if early domestic populations benefited from extensive gene flow from the wild is still to be revealed. It has been proposed that the high genetic similarity of modern domestic to Turkish wild emmer could be explained by a feralization of the very first proto-domestic population (Civáň et al. 2013; Oliveira et al. 2020). The analysis of wild and domestic samples from this region dating back to Pre-Pottery Neolithic and Neolithic could help determine the origin of the domestic pool, and its relationships with ancient and extant wild populations.
The recent genetic identification of domesticated T. timopheevii has triggered a re-evaluation of its importance and abundance in the archaeological record. This effort will be greatly aided by a genetic survey of the modern wild specimens, together with ancient seeds. In general, it will be interesting to use ancient and modern genetic data to compare the origins in space and time of parallel domestication events in wheat (emmer wheat, einkorn wheat, and T. timopheevii).
Prospects for the analysis of DNA from fully charred remains are poor, which limits the direct genetic analysis to unveil some of the earliest and most crucial events in wheat domestication. Nevertheless, we expect that improvements in the modelling of genomic evolution and the increasing availability of waterlogged remains will allow to test alternative scenarios on top of addressing questions concerning adaptation and spread of wheat.
7.5.2 Open Questions in Dispersal and Adaptation
The dispersal of wheat was accompanied by adaptation to different environments, leading to the evolutionary success of this species. An interesting example is adaptation to altitude along certain dispersal routes. Wild emmer wheat from the Northern Levant, the closest to all domestic landraces, is always found at high altitude. Its dispersal towards Egypt entailed cultivation at sea level, but emmer wheat grown on the Ethiopian plateau is cultivated at high altitudes again. There are two possible routes of dispersal leading to Ethiopia, one through Africa and another through the Iranian plateau and the Arabian Peninsula. The first one would entail a second adaptation event to high altitudes. The other would have always been cultivated at high altitudes, but there would require a longer dispersal route. How did emmer wheat arrive to Ethiopia? The analysis of desiccated specimens from the Arabian Peninsula, Sudan, and ideally Iran could help to answer this question, as well as potentially unveiling genetic mechanisms for adaptation to high altitude.
7.5.3 Open Questions in Hybridization and Speciation
Archaeological data increasingly suggests that different wheat species were used in a complex geographical mosaic that shifted through time. Given that several wheat species, i.e. emmer, einkorn, naked wheats, and T. timopheevi (and wild relatives) co-existed in the same area for millennia, we can ask how much genetic exchange was ongoing in Neolithic settlements. While the vast majority of wheat cultivated today is bread wheat, other free-threshing hexaploids such as the Indian dwarf wheat or the Yunan wheat could have arisen from different hybridization events, since the phylogeny of the A and B genomes differs from that of the D genome (Zhou et al. 2020). Furthermore, forms such as T. compactum (Club Wheat) have been described (e.g. Kaplan et al. 1992), even though it is unclear whether these morphotypes are the product of different hybridizations events or the consequence of differential selective pressures. A comparison of the D subgenome in ancient hexaploids with modern Aegilops specimens could tackle this question and narrow down the geographic origin where these hybridizations occurred.
Even more intriguingly, we can speculate whether introgressed genetic variation between different wheats was important for crop evolution and adaptation to different environments such as adaptation to northern latitudes or to heat stress. Einkorn wheat and spelt were important crops in central and northern Europe. On the other hand, hexaploid free-threshing wheats such as Indian dwarf wheat and T. compactum are more commonly found in warm environments. Studying changes in allele frequencies with the spread of these crops into new environments would identify candidate adaptive regions, whose phenotypic effects and usefulness could be analysed through crossing and genetic mapping. Learning from the phylogenetic relationship between ancient wheat specimens would greatly increase the power to detect the genomic regions conferring adaptation to those traits.
Furthermore, besides the impact that archaegenomics has on our understanding of the past, it has also the potential to set the basis for future food security (Pont et al. 2019b), conservation and breeding strategies, in the current context of climate change (di Donato et al. 2018). During the dispersal of domestic plants, crops adapted to a multitude of environments, and aDNA can reveal genetic diversity present in historical landraces but lost from the modern domestic pool (e.g. Scott et al. 2019). Detecting signals of positive selection in such lost diversity may therefore be particularly valuable, especially when it is the source of adaptations to extreme environments. After its identification, such diversity can be prioritized for preservation or introduced to modern cultivars via breeding if still present in seed banks, landraces, or wild relatives (di Donato et al. 2018). Plant aDNA studies can lead to the identification of lost crops and their wild relatives, revealing their genetic makeup. Such knowledge could set the ground for de novo domestications and ultimately aid in the diversification of our food system, which currently relies on a rather small number of domestic species (Estrada et al. 2018). Finally, aDNA can be informative of past plant-pathogens interactions and their co-evolution, e.g. (Yoshida et al. 2013), providing valuable insights for crop management (di Donato et al. 2018; Estrada et al. 2018; Przelomska et al. 2020).
In conclusion, archaeogenomics allows interrogation of a plethora of questions about wheat evolutionary history, such as population continuity and demographic changes through time, identification of climatic or cultural conditions that correspond to germplasm shifts, and relationships with other wheats. We expect these questions to be addressed in future aDNA studies. Overall, answering these questions will not only bring a deeper understanding of wheat evolution, but will also aid answering questions about human cultural evolution and trade.
References
Abbo S, Pinhasi van-Oss R, Gopher A, Saranga Y, Ofner I, Peleg Z (2014) Plant domestication versus crop evolution: a conceptual framework for cereals and grain legumes. Trends Plant Sci 19(6):351–360. https://doi.org/10.1016/j.tplants.2013.12.002
Allaby RG, Banerjee M, Brown TA (1999) Evolution of the high molecular weight glutenin loci of the A, B, D, and G genomes of wheat. Genome 42(2):296–307. https://doi.org/10.1139/g98-114
Alonge M, Shumate A, Puiu D, Zimin A, Salzberg SL (2020) Chromosome-scale assembly of the bread wheat genome reveals thousands of additional gene copies. Genetics. https://doi.org/10.1534/genetics.120.303501
Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, Pozniak CJ, Choulet F, Distelfeld A, Poland J, Ronen G, Barad O, Baruch K, Keeble-Gagnère G, Mascher M, Ben-Zvi G, Josselin AA, Himmelbach A, Balfourier F et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361(6403):1–163. https://doi.org/10.1126/science.aar7191
Arranz-Otaegui A, Colledge S, Zapata L, Teira-Mayolini LC, Ibáñez JJ (2016) Regional diversity on the timing for the initial appearance of cereal cultivation and domestication in southwest Asia. Proc Natl Acad Sci USA 113(49):14001–14006. https://doi.org/10.1073/pnas.1612797113
Arranz-Otaegui A, González Carretero L, Roe J, Richter T (2018) “Founder crops” v. wild plants: assessing the plant-based diet of the last hunter-gatherers in southwest Asia. Quatern Sci Rev 186:263–283. https://doi.org/10.1016/j.quascirev.2018.02.011
Ávila-Arcos MC, Cappellini E, Romero-Navarro JA, Wales N, Moreno-Mayar JV, Rasmussen M, Fordyce SL, Montiel R, Vielle-Calzada JP, Willerslev E, Gilbert MTP (2011) Application and comparison of large-scale solution-based DNA capture-enrichment methods on ancient DNA. Sci Rep 1. https://doi.org/10.1038/srep00074
Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale I, Mascher M, Spannagl M, Wiebe K, Jordan KW, Golan G, Deek J, Ben-zvi B, Ben-zvi G, Himmelbach A, Maclachlan RP, Sharpe AG, Komatsuda T et al (2017) Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 97(July):93–97. https://doi.org/10.1126/science.aan0032
Bilgic H, Hakki EE, Pandey A, Khan MK, Akkaya MS (2016) Ancient DNA from 8400 year-old Çatalhöyük wheat: implications for the origin of Neolithic agriculture. PLoS ONE 11(3):e0151974. https://doi.org/10.1371/journal.pone.0151974
Blatter RHE, Jacomet S, Schlumbaum A (2002) Spelt-specific alleles in HMW glutenin genes from modern and historical European spelt (Triticum spelta L.). Theor Appl Genet 104(2–3):329–337. https://doi.org/10.1007/s001220100680
Botigue LR, Song S, Scheu A, Gopalan S, Pendleton AL, Zeeb-lanz A, Arbogast R, Oetjens M, Taravella AM, Serege T, Burger J, Kidd JM, Veeramah KR, Bobo D, Daly K, Unterla M (2017) Ancient European dog genomes reveal continuity since the Early Neolithic. Nat Commun. https://doi.org/10.1038/ncomms16082
Briggs AW, Stenzel U, Johnson PLF, Pääbo S (2007) Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci 104:14616–14621. https://doi.org/10.1073/pnas.0704665104
Briggs AW, Stenzel U, Meyer M, Krause J, Kircher M, Pääbo S (2010) Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res 38. https://doi.org/10.1093/nar/gkp1163
Cheng H, Liu J, Wen J, Nie X, Xu L, Chen N, Li Z, Wang Q, Zheng Z, Li M, Cui L, Liu Z, Bian J, Wang Z, Xu S, Yang Q, Appels R, Han D, Song W et al (2019) Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol 20(1):1–16. https://doi.org/10.1186/s13059-019-1744-x
Civáň P, Ivaničová Z, Brown TA (2013) Reticulated origin of domesticated emmer wheat supports a dynamic model for the emergence of agriculture in the fertile crescent. PLoS ONE 8(11):1–11. https://doi.org/10.1371/journal.pone.0081955
Czajkowska BI, Bogaard A, Charles M, Jones G, Kohler-Schneider M, Mueller-Bieniek A, Brown TA (2020) Ancient DNA typing indicates that the “new” glume wheat of early Eurasian agriculture is a cultivated member of the Triticum timopheevii group. J Archaeol Sci 123(October):105258. https://doi.org/10.1016/j.jas.2020.105258
Da Fonseca RR, Smith BD, Wales N, Cappellini E, Skoglund P, Fumagalli M, Samaniego JA, Carøe C, Ávila-Arcos MC, Hufnagel DE, Korneliussen TS, Vieira FG, Jakobsson M, Arriaza B, Willerslev E, Nielsen R, Hufford MB, Albrechtsen A, Ross-Ibarra J, Gilbert MTP (2015) The origin and evolution of maize in the Southwestern United States. Nat Plants 1(January):1–5. https://doi.org/10.1038/nplants.2014.3
Daly KG, Delser PM, Mullin VE, Scheu A, Mattiangeli V, Teasdale MD, Hare AJ, Burger J, Verdugo MP, Collins MJ, Kehati R, Erek CM (2018) Ancient goat genomes reveal mosaic domestication in the Fertile Crescent. Science 361: 85–88, 24–27. https://doi.org/10.1126/science.aas9411
Der Sarkissian C, Allentoft ME, Avila-Arcos MC, Barnett R, Campos PF, Cappellini E, Ermini L, Fernandez R, da Fonseca R, Ginolhac A, Hansen AJ, Jonsson H, Korneliussen T, Margaryan A, Martin MD, Moreno-Mayar JV, Raghavan M, Rasmussen M, Velasco MS et al (2014) Ancient genomics. Philos Trans R Soc B: Biol Sci 370(1660):20130387–20130387. https://doi.org/10.1098/rstb.2013.0387
di Donato A, Filippone E, Ercolano MR, Frusciante L (2018) Genome sequencing of ancient plant remains: findings, uses and potential applications for the study and improvement of modern crops. Front Plant Sci 9(April). https://doi.org/10.3389/fpls.2018.00441
Diamond J (2002) Evolution, consequences and future of plant and animal domestication. Nature 418(6898):700–707. https://doi.org/10.1038/nature01019
Estrada O, Breen J, Richards SM, Cooper A (2018) Ancient plant DNA in the genomic era. Nat Plants 4(7):394–396. https://doi.org/10.1038/s41477-018-0187-9
Feldman M, Kislev EM (2007) Domestication of emmer wheat and evolution of free-threshing tetraploid wheat. Isr J Plant Sci 55:207–221
Fernández E, Thaw S, Brown TA, Arroyo-Pardo E, Buxò R, Serret MD, Araus JL (2013) DNA analysis in charred grains of naked wheat from several archaeological sites in Spain. J Archaeol Sci 659–670. https://doi.org/10.1016/j.jas.2012.07.014
Filipovic D (2014) Southwest Asian founder- and other crops at Neolithic sites in Serbia. Bul E-J Archaeol 4(2):229–247
Fornaciari R, Fornaciari S, Francia E, Mercuri AM, Arru L (2018) Panicum spikelets from the Early Holocene Takarkori rockshelter (SW Libya): Archaeo-molecular and -botanical investigations. Plant Biosyst 152(1):1–13. https://doi.org/10.1080/11263504.2016.1244117
Frantz LAF, Haile J, Lin AT, Scheu A, Benecke N, Alexander M, Linderholm A, Mullin VE, Daly KG, Battista VM, Price M, Gron KJ, Alexandri P, Arbogast R, Adrian B, Barnett R, Bartosiewicz L, Baryshnikov G, Bonsall C et al (2019) Ancient pigs reveal a near-complete genomic turnover following their introduction to Europe. Pnas 116. https://doi.org/10.1073/pnas.2008793117
Fuller DQ, Allaby RG, Stevens C (2010) Domestication as innovation: the entanglement of techniques, technology and chance in the domestication of cereal crops. World Archaeol 42(1):13–28. https://doi.org/10.1080/00438240903429680
Fuller DQ, Denham T, Arroyo-Kalin M, Lucas L, Stevens CJ, Qin L, Allaby RG, Purugganan MD (2014) Convergent evolution and parallelism in plant domestication revealed by an expanding archaeological record. Proc Natl Acad Sci USA 111(17):6147–6152. https://doi.org/10.1073/pnas.1308937110
Fulton TL (2012) Setting up an ancient DNA laboratory. In: Shapiro B, Hofreiter M (eds) Ancient DNA: methods and protocols. pp 1–11. https://doi.org/10.1007/978-1-61779-516-9_1
Gardiner LJ, Brabbs T, Akhunov A, Jordan K, Budak H, Richmond T, Singh S, Catchpole L, Akhunov E, Hall A (2019) Integrating genomic resources to present full gene and putative promoter capture probe sets for bread wheat. GigaScience 8(4):1–13. https://doi.org/10.1093/gigascience/giz018
Gaunitz C, Fages A, Hanghøj K, Albrechtsen A, Khan N, Schubert M, Seguin-orlando A, Owens IJ, Felkel S, Bignon-lau O, Damgaard PDB, Mittnik A, Wallner B, Merz V, Merz I, Zaibert V, Willerslev E (2018) Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. 1–5
Golenberg EM (1988) Outcrossing rates and their relationship to phenology in Triticum dicoccoides. Theoret Appl Genetics 75:937–944. https://doi.org/10.1007/BF00258057
Günther T, Nettelblad C (2019) The presence and impact of reference bias on population genomic studies of prehistoric human populations. PLoS Genet 15(7):1–20. https://doi.org/10.1371/journal.pgen.1008302
Gutaker RM, Burbano HA (2017) Reinforcing plant evolutionary genomics using ancient DNA. Curr Opin Plant Biol 36:38–45. https://doi.org/10.1016/j.pbi.2017.01.002
Gutaker RM, Weiß CL, Ellis D, Anglin NL, Knapp S, Fernández-alonso JL, Prat S, Burbano HA (2019) The origins and adaptation of European potatoes reconstructed from historical genomes. Nat Ecol Evol 3(July). https://doi.org/10.1038/s41559-019-0921-3
Haas M, Schreiber M, Mascher M (2018) Domestication and crop evolution of wheat and barley: genes, genomics, and future directions. J Integr Plant Biol XXXX(Xxxx). https://doi.org/10.1111/jipb.12737
He F, Pasam R, Shi F, Kant S, Keeble-Gagnere G, Kay P, Forrest K, Fritz A, Hucl P, Wiebe K, Knox R, Cuthbert R, Pozniak C, Akhunova A, Morrell PL, Davies JP, Webb SR, Spangenberg G, Hayes B et al (2019) Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat Genet 51(5):896–904. https://doi.org/10.1038/s41588-019-0382-2
Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC (1984) DNA sequences from the quagga, an extinct member of the horse family. Nature 312:282–284
Hofreiter M, Paijmans JLA, Goodchild H, Speller CF, Barlow A, Fortes GG, Thomas JA, Ludwig A, Collins MJ (2015) The future of ancient DNA: technical advances and conceptual shifts. BioEssays 37(3):284–293. https://doi.org/10.1002/bies.201400160
Jones G, Valamoti S, Charles M (2000) Early crop diversity: a “new” glume wheat from northern Greece. Veg Hist Archaeobot 9(3):133–146. https://doi.org/10.1007/BF01299798
Jonsson H, Korneliussen T, Margaryan A, Martin MD, Moreno-Mayar JV, Raghavan M, Rasmussen M, Velasco MS, Orlando L (2014) Ancient genomics. Phil Trans R Soc B 370. https://doi.org/10.1098/rstb.2013.0387
Jordan KW, Wang S, Lun Y, Gardiner LJ, MacLachlan R, Hucl P, Wiebe K, Wong D, Forrest KL, Sharpe AG, Sidebottom CHD, Hall N, Toomajian C, Close T, Dubcovsky J, Akhunova A, Talbert L, Bansal UK, Bariana HS et al (2015) A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biol 16(1):1–18. https://doi.org/10.1186/s13059-015-0606-4
Jorgensen C, Luo MC, Ramasamy R, Dawson M, Gill BS, Korol AB, Distelfeld A, Dvorak J (2017) A high-density genetic map of wild emmer wheat from the karaca dağ region provides new evidence on the structure and evolution of wheat chromosomes. Front Plant Sci 8(October):1–13. https://doi.org/10.3389/fpls.2017.01798
Kantar MB, Nashoba AR, Anderson JE, Blackman BK, Rieseberg LH (2017) The genetics and genomics of plant domestication. BioScience XX(X):1–12. https://doi.org/10.1093/biosci/bix114
Kaplan L, Smith MB, Sneddon LA (1992) Cereal grain phytoliths of Southwest Asia and Europe. In: Rapp G, Mulholland SC (eds) Phytolith systematics in archaeological and museum science. Springer US, pp 149–174. https://doi.org/10.1007/978-1-4899-1155-1_8
Key FM, Posth C, Esquivel-gomez LR, Hübler R, Spyrou MA, Neumann GU, Furtwängler A, Sabin S, Burri M, Wissgott A, Lankapalli AK, Vågene ÅJ, Meyer M, Nagel S, Tukhbatova R, Khokhlov A, Chizhevsky A, Hansen S, Belinsky AB et al (2020) Emergence of human-adapted Salmonella enterica is linked to the Neolithization process. Nat Ecol Evol 4(March). https://doi.org/10.1038/s41559-020-1106-9
Kistler L, Ware R, Smith O, Collins M, Allaby RG (2017) A new model for ancient DNA decay based on paleogenomic meta-analysis. Nucleic Acids Res 45:6310–6320. https://doi.org/10.1093/nar/gkx361
Kistler L, Bieker VC, Martin MD, Pedersen MW, Ramos Madrigal J, Wales N (2020) Ancient plant genomics in archaeology, herbaria, and the environment. Annu Rev Plant Biol 71:605–629. https://doi.org/10.1146/annurev-arplant-081519-035837
Korneliussen TS, Albrechtsen A, Nielsen R (2014) ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15:1–13. https://doi.org/10.1186/s12859-014-0356-4
Krause J, Fu Q, Good JM, Viola B, Shunkov MV, Derevianko AP (2010) The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature 464(April):894–897. https://doi.org/10.1038/nature08976
Lacan M, Keyser C, Ricaut F, Brucato N, Duranthon F, Guilaine J (2011) Ancient DNA reveals male diffusion through the Neolithic Mediterranean route.https://doi.org/10.1073/pnas.1100723108/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1100723108
Larson G, Piperno DR, Allaby RG, Purugganan MD, Andersson L, Arroyo-Kalin M, Barton L, Climer Vigueira C, Denham T, Dobney K et al (2014) Currrent perspectives and the future of domestication studies. Proc Natl Acad Sci 111(17):6139–6146. https://doi.org/10.1073/pnas.1323964111
Lawson DJ, van Dorp L, Falush D (2018) A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat Commun 9(1):1–11. https://doi.org/10.1038/s41467-018-05257-7
Leathlobhair MN, Perri AR, Irving-pease EK, Witt KE et al (2018) The evolutionary history of dogs in the Americas. 85(July):81–85
Lev-Yadun S, Gopher A, Abbo S (2000) The cradle of agriculture. Science 361. https://doi.org/10.1126/science.aao4776
Li C, Lister DL, Li H, Xu Y, Cui Y, Bower MA, Jones MK, Zhou H (2011) Ancient DNA analysis of desiccated wheat grains excavated from a Bronze Age cemetery in Xinjiang. J Archaeol Sci 38(1):115–119. https://doi.org/10.1016/j.jas.2010.08.016
Librado P, Khan N, Fages A et al (2021) The origins and spread of domestic horses from the Western Eurasian steppes. Nature 598. https://doi.org/10.1038/s41586-021-04018-9
Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362:709–715. https://doi.org/10.1038/362709a0
Link V, Kousathanas A, Veeramah K, Sell C, Scheu A, Wegmann D (2017) ATLAS: analysis tools for low-depth and ancient samples. BioRxiv 33(16):1–7
Ling HQ, Ma B, Shi X, Liu H, Dong L, Sun H, Cao Y, Gao Q, Zheng S, Li Y, Yu Y, Du H, Qi M, Li Y, Lu H, Yu H, Cui Y, Wang N, Chen C et al (2018) Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557(7705):424–428. https://doi.org/10.1038/s41586-018-0108-0
Luo MC, Yang ZL, You FM, Kawahara T, Waines JG, Dvorak J (2007) The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor Appl Genet 114(6):947–959. https://doi.org/10.1007/s00122-006-0474-0
Luo MC, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, Huo N, Zhu T, Wang L, Wang Y, McGuire PE, Liu S, Long H, Ramasamy RK, Rodriguez JC, Van Sonny L, Yuan L, Wang Z, Xia Z et al (2017) Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551(7681):498–502. https://doi.org/10.1038/nature24486
Maccaferri M, Harris NS, Twardziok SO, Pasam RK, Gundlach H, Spannagl M, Ormanbekova D, Lux T, Prade VM, Milner SG, Himmelbach A, Mascher M, Bagnaresi P, Faccioli P, Cozzi P, Lauria M, Lazzari B, Stella A, Manconi A et al (2019) Durum wheat genome highlights past domestication signatures and future improvement targets. Nat Genet 51(5):885–895.https://doi.org/10.1038/s41588-019-0381-3
Marciniak S, Perry GH (2017) Harnessing ancient genomes to study the history of human adaptation. Nat Publ Group 18(11):659–674. https://doi.org/10.1038/nrg.2017.65
Mascher M, Schuenemann VJ, Davidovich U, Marom N, Himmelbach A, Hübner S, Korol A, David M, Reiter E, Riehl S, Schreiber M, Vohr SH, Green RE, Dawson IK, Russell J, Kilian B, Muehlbauer GJ, Waugh R, Fahima T et al (2016). Genomic analysis of 6000-year-old cultivated grain illuminates the domestication history of barley. Nat Genet 48(9):1089–1093. https://doi.org/10.1038/ng.3611
McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Ripatti S (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48:1279–1283
Morozova I, Flegontov P, Mikheyev AS, Bruskin S, Asgharian H, Ponomarenko P, Klyuchnikov V, Kumar GPA, Prokhortchouk E, Gankin Y, Rogaev E, Nikolsky Y, Baranova A, Elhaik E, Tatarinova TV (2016) Toward high-resolution population genomics using archaeological samples. DNA Res 23(4):295–310. https://doi.org/10.1093/dnares/dsw029
Nave M, Avni R, Çakır E, Portnoy V, Sela H, Pourkheirandish M, Ozkan H, Hale I, Komatsuda T, Dvorak J, Distelfeld A (2019) Wheat domestication in light of haplotype analyses of the Brittle rachis 1 genes (BTR1-A and BTR1-B). Plant Sci 285(May):193–199. https://doi.org/10.1016/j.plantsci.2019.05.012
Nesbitt M, Samuel D (1996) From staple crop to extinction? The archaeology and history of the hulled wheat. In: Padulosi S, Hammer K, Heller J (eds) Hulled wheats, promoting the conservation and used of underutilized and neglected crops. pp 40–99
Nielsen R, Akey JM, Jakobsson M, Pritchard JK, Tishkoff S, Willerslev E (2017) Tracing the peopling of the world through genomics. Nature 541. https://doi.org/10.1038/nature21347
Oliveira HR, Jacocks L, Czajkowska BI, Kennedy SL, Brown TA (2020) Multiregional origins of the domesticated tetraploid wheats. PLoS ONE 15(1):1–20. https://doi.org/10.1371/journal.pone.0227148
Orlando L, Allaby R, Pontus S, Der Sarkissian C, Stockhammer PW, Ávila-Arcos MC, Fu Q, Krause J, Willerslev E, Stone A, Christina W (2021) Ancient DNA analysis. Nat Rev Genet 1–26. https://doi.org/10.1038/s43586-020-00011-0
Özbaşaran M, Duru G, Stiner MC, Esin U (2018) The early settlement at Aşıklı Höyük: essays in honor of Ufuk Esin. Ege Yayınları
Ozkan H, Brandolini A, Schafer-Pregl R, Salamini F (2002) AFLP analysis of a collection of tetraploid wheat indicated the origin of emmer and hard wheat domestication in south-eastern Turkey. Mol Biol Evol 19:1797–1801
Ozkan H, Brandolini A, Pozzi C, Effgen S, Wunder J, Salamini F (2005) A reconsideration of the domestication geography of tetraploid wheats. Theor Appl Genet 110:1052–1060
Palmer SA, Moore JD, Clapham AJ, Rose P, Allaby RG (2009) Archaeogenetic evidence of ancient nubian barley evolution from six to two-row indicates local adaptation. PLoS ONE 4(7):2–8. https://doi.org/10.1371/journal.pone.0006301
Palmer SA, Clapham AJ, Rose P, Freitas FO, Owen BD, Beresford-Jones D, Moore JD, Kitchen JL, Allaby RG (2012) Archaeogenomic evidence of punctuated genome evolution in gossypium. Mol Biol Evol 29(8):2031–2038. https://doi.org/10.1093/molbev/mss070
Parducci L, Bennett KD, Ficetola GF, Alsos IG, Suyama Y, Wood JR, Pedersen MW (2017) Ancient plant DNA in lake sediments. New Phytol 214(3):924–942. https://doi.org/10.1111/nph.14470
Pedersen JS, Valen E, Velazquez AMV, Parker BJ, Rasmussen M, Lindgreen S, Lilje B, Tobin DJ, Kelly TK, Vang S, Andersson R, Jones PA, Hoover CA, Tikhonov A, Prokhortchouk E, Rubin EM, Sandelin A, Gilbert MTP, Krogh A et al (2014) Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome 454–466. https://doi.org/10.1101/gr.163592.113.Freely
Peltzer A, Jäger G, Herbig A, Seitz A, Kniep C, Krause J, Nieselt K (2016) EAGER: efficient ancient genome reconstruction. Genome Biol 17:60. https://doi.org/10.1186/s13059-016-0918-z
Peyrégne S, Prüfer K (2020) Present-day DNA contamination in ancient DNA datasets. BioEssays 42(1–11). https://doi.org/10.1002/bies.202000081
Pont C, Leroy T, Seidel M, Tondelli A, Duchemin W, Armisen D, Lang D, Bustos-Korts D, Goué N, Balfourier F, Molnár-Láng M, Lage J, Kilian B, Özkan H, Waite D, Dyer S, Letellier T, Alaux M, Russell J et al (2019a) Tracing the ancestry of modern bread wheats. Nat Genet 51(5):905–911. https://doi.org/10.1038/s41588-019-0393-z
Pont C, Wagner S, Kremer A, Orlando L, Plomion C, Salse J (2019b) Paleogenomics: reconstruction of plant evolutionary trajectories from modern and ancient DNA. Genome Biol 20(1):29. https://doi.org/10.1186/s13059-019-1627-1
Prüfer K, Meyer M (2015) Comment on “Late Pleistocene human skeleton and mtDNA link Paleoamericans and modern Native Americans.” Science 347(6224):835. https://doi.org/10.1126/science.1260617
Przelomska NAS, Armstrong CG, Kistler L (2020) Ancient plant DNA as a window into the cultural heritage and biodiversity of our food system. Front Ecol Evol 8(March):1–8. https://doi.org/10.3389/fevo.2020.00074
Przewieslik-Allen AM, Wilkinson PA, Burridge AJ, Winfield MO, Dai X, Beaumont M, King J, Yang C, Griffiths S, Wingen LU, Horsnell R, Bentley AR, Shewry P, Barker GLA, Edwards KJ (2021) The role of gene flow and chromosomal instability in shaping the bread wheat genome. Nat Plants 7(2):172–183. https://doi.org/10.1038/s41477-020-00845-2
Purugganan MD, Fuller DQ (2009) The nature of selection during plant domestication. Nature 457(7231):843–848. https://doi.org/10.1038/nature07895
Ramos-Madrigal J, Smith BD, Moreno-Mayar JV, Gopalakrishnan S, Ross-Ibarra J, Gilbert MTP, Wales N (2016) Genome sequence of a 5310-year-old maize cob provides insights into the early stages of maize domestication. Curr Biol 26(23):3195–3201. https://doi.org/10.1016/j.cub.2016.09.036
Ramos-Madrigal J, Runge AKW, Bouby L, Lacombe T, Samaniego Castruita JA, Adam-Blondon AF, Figueiral I, Hallavant C, Martínez-Zapater JM, Schaal C, Töpfer R, Petersen B, Sicheritz-Pontén T, This P, Bacilieri R, Gilbert MTP, Wales N (2019) Palaeogenomic insights into the origins of French grapevine diversity. Nat Plants 5(6):595–603. https://doi.org/10.1038/s41477-019-0437-5
Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, Maricic T, Good JM, Marques-bonet T, Alkan C, Fu Q, Mallick S, Li H, Meyer M, Eichler EE, Stoneking M (2010) Genetic history of an archaic hominin group from Denisova Cave in Siberia. https://doi.org/10.1038/nature09710
Renaud G, Schubert M, Sawyer S, Orlando L (2019) Authentication and assessment of contamination in ancient DNA. In: Shapiro B et al (eds) Ancient DNA: methods and protocols. Springer US, pp 163–194. https://doi.org/10.2307/j.ctt183pd9z.20
Renner SS, Pérez-Escbar OA, Silber MV, Nesbitt M, Preick M, Hofreiter M, Chomicki G (2019) A 3500-year-old leaf from a Pharaonic tomb reveals that New Kingdom Egyptians were cultivating domesticated watermelon. BioRxiv. https://doi.org/10.1101/642785
Rohland N, Harney E, Mallick S, Nordenfelt S, Reich D (2015) Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Phil Trans R Soc B 370(1660). https://doi.org/10.1098/rstb.2013.0624
Schubert M, Ermini L, Sarkissian CD, Jónsson H, Ginolhac A, Schaefer R, Martin MD, Fernández R, Kircher M, McCue M, Willerslev E, Orlando L (2014) Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat Protoc 9(5):1056–1082. https://doi.org/10.1038/nprot.2014.063
Scott MF, Botigué LR, Brace S, Stevens CJ, Mullin VE, Stevenson A, Thomas MG, Fuller DQ, Mott R (2019) A 3000-year-old Egyptian emmer wheat genome reveals dispersal and domestication history. Nat Plants 5(11):1120–1128. https://doi.org/10.1038/s41477-019-0534-5
Sharma JS, Running KLD, Xu SS, Zhang Q, Peters Haugrud AR, Sharma S, McClean PE, Faris JD (2019) Genetic analysis of threshability and other spike traits in the evolution of cultivated emmer to fully domesticated durum wheat. Mol Genet Genomics 294(3):757–771. https://doi.org/10.1007/s00438-019-01544-0
Smith O, Nicholson WV, Kistler L, Mace E, Clapham A, Rose P, Stevens C, Ware R, Samavedam S, Barker G, Jordan D, Fuller DQ, Allaby RG (2019) A domestication history of dynamic adaptation and genomic deterioration in Sorghum. Nat Plants 5(April). https://doi.org/10.1038/s41477-019-0397-9
Snir A, Nadel D, Groman-Yaroslavski I, Melamed Y, Sternberg M, Bar-Yosef O, Weiss E (2015) The origin of cultivation and proto-weeds, long before neolithic farming. PLoS ONE 10(7):1–12. https://doi.org/10.1371/journal.pone.0131422
Spyrou MA, Bos KI, Herbig A, Krause J (2019) Ancient pathogen genomics as an emerging tool for infectious disease research. Nat Rev Genet 20(June). https://doi.org/10.1038/s41576-019-0119-1
Swarts K, Gutaker RM, Benz B, Blake M, Bukowski R, Holland J, Kruse-peeples M, Lepak N, Prim L, Romay MC, Ross-ibarra J, Sanchez-gonzalez JDJ (2017) Genomic estimation of complex traits reveals ancient maize adaptation to temperate North America. 515(August):512–515. https://doi.org/10.1126/science.aam9425Estimating
Trucchi E, Benazzo A, Lari M, Iob A, Vai S, Nanni L, Bellucci E, Bitocchi E, Raffini F, Xu C, Jackson SA, Lema V, Babot P, Oliszewski N, Gil A, Neme G, Michieli CT, De Lorenzi M, Calcagnile L et al (2021) Ancient genomes reveal early Andean farmers selected common beans while preserving diversity. Nat Plants 7(2):123–128. https://doi.org/10.1038/s41477-021-00848-7
Ulaş B, Fiorentino G (2021) Recent attestations of “new ” glume wheat in Turkey: a reassessment of its role in the reconstruction of Neolithic agriculture. Veg Hist Archaeobotany 30(5):685–701. https://doi.org/10.1007/s00334-020-00807-w
Vavilov NI, Vavylov MI, Vavílov NÍ, Dorofeev VF (1992) Origin and geography of cultivated plants. Cambridge University of Press
Verdugo MP, Mullin VE, Scheu A, Mattiangeli V, Daly KG, Delser PM, Hare AJ, Burger J, Collins MJ, Kehati R, Hesse P, Fulton D (2019) Ancient cattle genomics, origins, and rapid turnover in the Fertile Crescent. 176(July):173–176. https://doi.org/10.5281/zenodo.3206663
Wagenaar EB (1966) Studies on the genome constitution of Triticum timopheevi Zhuk. II. The T. timopheevi complex and its origin. Soc Study Evol 20(2):150–164. https://www.jstor.org/stable/2406569
Wales N, Akman M, Watson RHB, Sánchez Barreiro F, Smith BD, Gremillion KJ, Gilbert MTP, Blackman BK (2018) Ancient DNA reveals the timing and persistence of organellar genetic bottlenecks over 3000 years of sunflower domestication and improvement. Evol Appl 12(December 2017):1–16. https://doi.org/10.1111/eva.12594
Wales N, Akman M, Watson RHB, Sánchez Barreiro F, Smith BD, Gremillion KJ, Gilbert MTP, Blackman BK (2019) Ancient DNA reveals the timing and persistence of organellar genetic bottlenecks over 3000 years of sunflower domestication and improvement. Evol Appl 12(1):38–53. https://doi.org/10.1111/eva.12594
Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D, Klymiuk V, Byrns B, Gundlach H, Bandi V, Siri JN, Nilsen K, Aquino C, Himmelbach A, Copetti D et al (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature 588(7837):277–283. https://doi.org/10.1038/s41586-020-2961-x
Warinner C, Rodrigues JFM, Vyas R, Trachsel C, Shved N, Grossmann J, Radini A, Hancock Y, Tito RY, Fiddyment S, Speller C, Hendy J, Charlton S, Luder HU, Salazar-garcía DC, Eppler E, Seiler R, Hansen LH, Alfredo J et al (2014) Pathogens and host immunity in the ancient human oral cavity. Nat Genet 46(4). https://doi.org/10.1038/ng.2906
Weyrich LS, Dobney K, Cooper A (2015) Ancient DNA analysis of dental calculus. J Hum Evol 79:119–124. https://doi.org/10.1016/j.jhevol.2014.06.018
Wu X, Ding B, Zhang B, Feng J, Wang Y, Ning C, Wu H, Zhang F, Zhang Q, Li N, Zhang Z, Sun X, Zhang Q, Li W, Liu B, Cui Y, Gong L (2019) Phylogenetic and population structural inference from genomic ancestry maintained in present-day common wheat Chinese landraces. Plant J 99(2):201–215. https://doi.org/10.1111/tpj.14421
Yang MA, Fan X, Sun B, Chen C, Lang J, Ko Y, Tsang C, Chiu H, Wang T, Bao Q, Wu X, Hajdinjak M, Ko AM, Ding M, Cao P, Yang R, Liu F, Nickel B, Dai Q et al (2020) Ancient DNA indicates human population shifts and admixture in northern and southern China. Science 288(July):282–288. https://doi.org/10.1126/science.aba0909
Yoshida K, Schuenemann VJ, Cano LM, Pais M, Mishra B, Sharma R et al (2013) The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. Elife 2:1–25. https://doi.org/10.7554/eLife.00731.001
Zhou Y, Zhao X, Li Y, Xu J, Bi A, Kang L, Xu D, Chen H, Wang Y, Wang Y-G, Liu S, Jiao C, Lu H, Wang J, Yin C, Jiao Y, Lu F (2020) Triticum population sequencing provides insights into wheat adaptation. Nat Genet 52(12):1412–1422. https://doi.org/10.1038/s41588-020-00722-w
Zohary D (2013) Domestication of Crop Plants. In: Encyclopedia of biodiversity, 2nd edn. pp 657–664. https://doi.org/10.1016/B978-0-12-384719-5.00199-4
Zhu T, Wang L, Rodriguez JC, Deal KR, Avni R, Distelfeld A, McGuire PE, Dvorak J, Luo MC (2019) Improved genome sequence of wild emmer wheat Zavitan with the aid of optical maps. G3: Genes, Genomes, Genet 9(3):619–624. https://doi.org/10.1534/g3.118.200902
Acknowledgements
A. I. is a FPI fellow (PRE2018-083529). L. B. is a Ramón y Cajal Fellow. (RYC2018-024770-I) both fellowships funded by the Ministerio de Ciencia e Innovación—Agencia Estatal de Investigación/ Fondo Social Europeo. We acknowledge financial support from the Spanish Agencia Estatal de Investigación (Ministry of Science and Innovation-State Research Agency) (AEI), through the “Severo Ochoa Programme for Centres of Excellence in R&D” SEV‐2015‐0533 and CEX2019-000902-S. This work was also supported by the CERCA programme by the Generalitat de Catalunya.
M. F. S. is supported by a Leverhulme Trust Early Career Fellowship (ECF-2020-095).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2024 The Author(s)
About this chapter
Cite this chapter
Iob, A., Scott, M.F., Botigué, L. (2024). Ancient Wheat Genomes Illuminate Domestication, Dispersal, and Diversity. In: Appels, R., Eversole, K., Feuillet, C., Gallagher, D. (eds) The Wheat Genome. Compendium of Plant Genomes. Springer, Cham. https://doi.org/10.1007/978-3-031-38294-9_7
Download citation
DOI: https://doi.org/10.1007/978-3-031-38294-9_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-38292-5
Online ISBN: 978-3-031-38294-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)