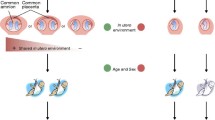
Abstract
Twin studies have played an important role in our understanding of individual variation for over a century. The strength of these lies in the capacity to almost perfectly control for inter-individual genetic variation through the study of monozygotic (MZ) twin pairs. Despite their genetic identity, MZ twins usually show phenotypic variability, often solely ascribed to non-shared environmental exposures. Given that epigenetic mechanisms are widely believed to be the mediators of the influence of environmental factors on the underlying genome, it is not surprising that study of twins in epigenetic research is an important approach to help unravel the complexities associated with gene: environment interactions in human development and disease. In addition, the strategic use of twins in epigenetic studies has revealed the importance of genetic factors and both in utero and postnatal environments to the establishment and maintenance of the human epigenome. However, a note of caution is warranted given emerging evidence for epigenetic variation as an inherent feature of MZ twinning and the potential for MZ twins to show genetic variability. Irrespective of this, twin studies are beginning to reveal evidence linking epigenetic disruption to disease-associated risk in humans.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- 2D:
-
Two dimensional
- 3′UTR:
-
Non-protein coding 3′ untranslated region of RNA transcript
- 5MeC:
-
5-methylcytosine
- AIMS:
-
Amplification of inter-methylated sites
- ANOVA:
-
Analysis of variance
- CBMC:
-
Cord blood mononuclear cells
- CGI:
-
CpG island region (enriched for CpG dinucleotides)
- COMT:
-
Catechol-O-methyltransferase
- DC:
-
Dichorionic (two placentas)
- DM:
-
Dermatomyositis
- DMR:
-
Differentially methylated regions
- DNA:
-
Deoxyribonucleic acid
- DRD2:
-
Dopaminergic receptor D2
- DZ:
-
Dizygotic
- EEA:
-
Equal environments assumption
- HUVEC:
-
Human umbilical vein endothelial cells
- ICC:
-
Intraclass correlation
- MC:
-
Monochorionic
- MSRDA:
-
Methylation-sensitive representational difference analysis
- MZ:
-
Monozygotic
- PPIEL:
-
Peptidylprolyl isomerase E-like
- RA:
-
Rheumatoid arthritis
- RNA:
-
Ribonucleic acid
- RRBS:
-
Reduced representation bisulphite sequencing
- SLE:
-
Systemic lupus erythematosus
- SMS:
-
Spermine synthase
References
Hasler R et al (2009) Genetic control of global gene expression levels in the intestinal mucosa: a human twin study. Physiol Genomics 38(1):73–79
Sharma K (2005) Genetic epidemiology of epilepsy: a twin study. Neurol India 53(1):93–98
Tan Q et al (2005) Genetic dissection of gene expression observed in whole blood samples of elderly Danish twins. Hum Genet 117(2–3):267–274
Gordon L et al (2012) Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res 22(8):1395–1406
Gordon L et al (2011) Clues to the effect of intrauterine milieu on gene expression from a study of two tissues from neonatal monozygotic twins. Epigenetics 6(5):579–592
Gartner K (1990) A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Lab Anim 24(1):71–77
Kerkel K et al (2008) Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 40(7):904–908
Schalkwyk LC et al (2010) Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet 86(2):196–212
Fuke C et al (2004) Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet 68(Pt 3):196–204
Fraga MF et al (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 102(30):10604–10609
Ollikainen M et al (2010) DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet 19(21):4176–4188
Tan Q et al (2015) Twin methodology in epigenetic studies. J Exp Biol 218(Pt 1):134–139
Boomsma D, Busjahn A, Peltonen L (2002) Classical twin studies and beyond. Nat Rev Genet 3(11):872–882
Bell JT, Spector TD (2011) A twin approach to unraveling epigenetics. Trends Genet 27:116–125
Heijmans BT et al (2007) Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet 16(5):547–554
Kaminsky ZA et al (2009) DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 41(2):240–245
McRae AF et al (2014) Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol 15(5):R73
Villicana S, Bell JT (2021) Genetic impacts on DNA methylation: research findings and future perspectives. Genome Biol 22(1):127
Min JL et al (2021) Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet 53(9):1311–1321
van Dongen J et al (2021) Identical twins carry a persistent epigenetic signature of early genome programming. Nat Commun 12(1):5618
Wong CC et al (2010) A longitudinal study of epigenetic variation in twins. Epigenetics 5(6):516–526
Spector TD (2000) Advances in twin and sib-pair analysis. Greenwich Medical Media
Martino D et al (2013) Longitudinal, genome-scale analysis of DNA methylation in twins from birth to 18 months of age reveals rapid epigenetic change in early life and pair-specific effects of discordance. Genome Biol 14(5):R42
Levesque ML et al (2014) Genome-wide DNA methylation variability in adolescent monozygotic twins followed since birth. Epigenetics 9(10):1410–1421
Horvath S (2013) DNA methylation age of human tissues and cell types. Genome Biol 14(10):R115
Knight AK et al (2016) An epigenetic clock for gestational age at birth based on blood methylation data. Genome Biol 17(1):206
Lee Y et al (2019) Placental epigenetic clocks: estimating gestational age using placental DNA methylation levels. Aging (Albany NY) 11(12):4238–4253
Girchenko P et al (2017) Associations between maternal risk factors of adverse pregnancy and birth outcomes and the offspring epigenetic clock of gestational age at birth. Clin Epigenetics 9:49
McCartney DL et al (2021) Genome-wide association studies identify 137 genetic loci for DNA methylation biomarkers of aging. Genome Biol 22(1):194
Reynolds CA et al (2020) A decade of epigenetic change in aging twins: genetic and environmental contributions to longitudinal DNA methylation. Aging Cell 19(8):e13197
Jylhava J et al (2019) Longitudinal changes in the genetic and environmental influences on the epigenetic clocks across old age: evidence from two twin cohorts. EBioMedicine 40:710–716
Fohr T et al (2021) Does the epigenetic clock GrimAge predict mortality independent of genetic influences: an 18 year follow-up study in older female twin pairs. Clin Epigenetics 13(1):128
Li S et al (2020) Genetic and environmental causes of variation in epigenetic aging across the lifespan. Clin Epigenetics 12(1):158
Zwijnenburg PJ, Meijers-Heijboer H, Boomsma DI (2010) Identical but not the same: the value of discordant monozygotic twins in genetic research. Am J Med Genet B Neuropsychiatr Genet 153B(6):1134–1149
Sakuntabhai A et al (1999) Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nat Genet 21(3):271–277
Robertson SP et al (2006) Postzygotic mutation and germline mosaicism in the otopalatodigital syndrome spectrum disorders. Eur J Hum Genet 14(5):549–554
Kondo S et al (2002) Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet 32(2):285–289
Smith AC et al (2006) New chromosome 11p15 epigenotypes identified in male monozygotic twins with Beckwith-Wiedemann syndrome. Cytogenet Genome Res 113(1–4):313–317
Helderman-van den Enden AT et al (1999) Monozygotic twin brothers with the fragile X syndrome: different CGG repeats and different mental capacities. J Med Genet 36(3):253–257
Kruyer H et al (1994) Fragile X syndrome and the (CGG)n mutation: two families with discordant MZ twins. Am J Hum Genet 54(3):437–442
Bonilla E et al (1990) Partial dystrophin deficiency in monozygous twin carriers of the Duchenne gene discordant for clinical myopathy. Neurology 40(8):1267–1270
Biousse V et al (1997) De novo 14484 mitochondrial DNA mutation in monozygotic twins discordant for Leber’s hereditary optic neuropathy. Neurology 49(4):1136–1138
Blakely EL et al (2004) Mitochondrial DNA deletion in “identical” twin brothers. J Med Genet 41(2):e19
Bruder CE et al (2008) Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet 82(3):763–771
Bakaysa SL et al (2007) Telomere length predicts survival independent of genetic influences. Aging Cell 6(6):769–774
Slagboom PE, Droog S, Boomsma DI (1994) Genetic determination of telomere size in humans: a twin study of three age groups. Am J Hum Genet 55(5):876–882
Jonsson H et al (2021) Differences between germline genomes of monozygotic twins. Nat Genet 53(1):27–34
Petronis A et al (2003) Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr Bull 29(1):169–178
Mill J et al (2006) Evidence for monozygotic twin (MZ) discordance in methylation level at two CpG sites in the promoter region of the catechol-O-methyltransferase (COMT) gene. Am J Med Genet B Neuropsychiatr Genet 141B(4):421–425
Mitchell MM et al (2011) Epigenetic investigation of variably X chromosome inactivated genes in monozygotic female twins discordant for primary biliary cirrhosis. Epigenetics 6(1):95–102
Weksberg R et al (2002) Discordant KCNQ1OT1 imprinting in sets of monozygotic twins discordant for Beckwith-Wiedemann syndrome. Hum Mol Genet 11(11):1317–1325
Yamazawa K et al (2008) Monozygotic female twins discordant for silver-Russell syndrome and hypomethylation of the H19-DMR. J Hum Genet 53(10):950–955
Oates NA et al (2006) Increased DNA methylation at the AXIN1 gene in a monozygotic twin from a pair discordant for a caudal duplication anomaly. Am J Hum Genet 79(1):155–162
Mastroeni D et al (2009) Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PLoS One 4(8):e6617
Souren NY et al (2011) DNA methylation variability at growth-related imprints does not contribute to overweight in monozygotic twins discordant for BMI. Obesity (Silver Spring) 19:1519–1522
Javierre BM et al (2010) Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res 20(2):170–179
Baranzini SE et al (2010) Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature 464(7293):1351–1356
Kuratomi G et al (2008) Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Mol Psychiatry 13(4):429–441
Kaminsky Z et al (2008) Epigenetics of personality traits: an illustrative study of identical twins discordant for risk-taking behavior. Twin Res Hum Genet 11(1):1–11
Yuan W et al (2014) An integrated epigenomic analysis for type 2 diabetes susceptibility loci in monozygotic twins. Nat Commun 5:5719
Carry PM et al (2021) Severity of idiopathic scoliosis is associated with differential methylation: an epigenome-wide association study of monozygotic twins with idiopathic scoliosis. Genes (Basel) 12(8):1191
Wang W et al (2021) Genome-wide DNA methylation and gene expression analyses in monozygotic twins identify potential biomarkers of depression. Transl Psychiatry 11(1):416
Paul DS et al (2016) Increased DNA methylation variability in type 1 diabetes across three immune effector cell types. Nat Commun 7:13555
Nilsson E et al (2014) Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 63(9):2962–2976
Wei M et al (2021) Monozygotic twins discordant for immunoglobulin a nephropathy display differences in DNA methylation and gene expression. Kidney Dis (Basel) 7(3):200–209
Young JI et al (2021) DNA methylation variation is identified in monozygotic twins discordant for non-syndromic cleft lip and palate. Front Cell Dev Biol 9:656865
Grunert M et al (2020) The needle in the haystack-searching for genetic and epigenetic differences in monozygotic twins discordant for tetralogy of Fallot. J Cardiovasc Dev Dis 7(4):55
Roberson-Nay R et al (2020) An epigenome-wide association study of early-onset major depression in monozygotic twins. Transl Psychiatry 10(1):301
Zhu Y et al (2019) Genome-wide profiling of DNA methylome and transcriptome in peripheral blood monocytes for major depression: a monozygotic discordant twin study. Transl Psychiatry 9(1):215
Liang S et al (2019) Genome-wide DNA methylation analysis reveals epigenetic pattern of SH2B1 in Chinese monozygotic twins discordant for autism Spectrum disorder. Front Neurosci 13:712
Mohandas N et al (2019) Evidence for type-specific DNA methylation patterns in epilepsy: a discordant monozygotic twin approach. Epigenomics 11(8):951–968
Souren NY et al (2019) DNA methylation signatures of monozygotic twins clinically discordant for multiple sclerosis. Nat Commun 10(1):2094
Webster AP et al (2018) Increased DNA methylation variability in rheumatoid arthritis-discordant monozygotic twins. Genome Med 10(1):64
Murphy TM et al (2015) Methylomic markers of persistent childhood asthma: a longitudinal study of asthma-discordant monozygotic twins. Clin Epigenetics 7:130
Ollikainen M et al (2015) Genome-wide blood DNA methylation alterations at regulatory elements and heterochromatic regions in monozygotic twins discordant for obesity and liver fat. Clin Epigenetics 7:39
Cobos I et al (2005) Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci 8(8):1059–1068
Wu TD, Nacu S (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26(7):873–881
Bennett CM, Boye E, Neufeld EJ (2008) Female monozygotic twins discordant for hemophilia a due to nonrandom X-chromosome inactivation. Am J Hematol 83(10):778–780
Willemsen R et al (2000) Twin sisters, monozygotic with the fragile X mutation, but with a different phenotype. J Med Genet 37(8):603–604
Zneimer SM, Schneider NR, Richards CS (1993) In situ hybridization shows direct evidence of skewed X inactivation in one of monozygotic twin females manifesting Duchenne muscular dystrophy. Am J Med Genet 45(5):601–605
Shur N (2009) The genetics of twinning: from splitting eggs to breaking paradigms. Am J Med Genet C Semin Med Genet 151C(2):105–109
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Saffery, R., Bell, J.T. (2022). The Utility of Twins for Epigenetic Analysis. In: Michels, K.B. (eds) Epigenetic Epidemiology. Springer, Cham. https://doi.org/10.1007/978-3-030-94475-9_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-94475-9_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-94474-2
Online ISBN: 978-3-030-94475-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)