Abstract
The 26S proteasome is the most complex ATP-dependent protease machinery, of ~2.5 MDa mass, ubiquitously found in all eukaryotes. It selectively degrades ubiquitin-conjugated proteins and plays fundamentally indispensable roles in regulating almost all major aspects of cellular activities. To serve as the sole terminal “processor” for myriad ubiquitylation pathways, the proteasome evolved exceptional adaptability in dynamically organizing a large network of proteins, including ubiquitin receptors, shuttle factors, deubiquitinases, AAA-ATPase unfoldases, and ubiquitin ligases, to enable substrate selectivity and processing efficiency and to achieve regulation precision of a vast diversity of substrates. The inner working of the 26S proteasome is among the most sophisticated, enigmatic mechanisms of enzyme machinery in eukaryotic cells. Recent breakthroughs in three-dimensional atomic-level visualization of the 26S proteasome dynamics during polyubiquitylated substrate degradation elucidated an extensively detailed picture of its functional mechanisms, owing to progressive methodological advances associated with cryogenic electron microscopy (cryo-EM). Multiple sites of ubiquitin binding in the proteasome revealed a canonical mode of ubiquitin-dependent substrate engagement. The proteasome conformation in the act of substrate deubiquitylation provided insights into how the deubiquitylating activity of RPN11 is enhanced in the holoenzyme and is coupled to substrate translocation. Intriguingly, three principal modes of coordinated ATP hydrolysis in the heterohexameric AAA-ATPase motor were discovered to regulate intermediate functional steps of the proteasome, including ubiquitin-substrate engagement, deubiquitylation, initiation of substrate translocation and processive substrate degradation. The atomic dissection of the innermost working of the 26S proteasome opens up a new era in our understanding of the ubiquitin-proteasome system and has far-reaching implications in health and disease.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Proteasome
- Ubiquitylation
- Ubiquitin-proteasome system
- Deubiquitylation
- AAA-ATPase motor
- Proteolysis
- Mechanochemistry
- Homeostasis
- Cryogenic electron microscopy
- Cryo-EM
- Conformational dynamics
Introduction
Human cells express more than 20,000 genes, which direct synthesis of a comparable number of proteins by the ribosome. Intracellular constituents dynamically preserved as proteins are continuously synthesized and degraded in cells. Newly synthesized proteins might misfold into dysfunctional structures that are cytotoxic and must be removed in a timely manner. Many initially well-folded proteins may need to be partially degraded or cleaved to activate specific function. Functional proteins may ultimately become unnecessary once they fulfil their missions. All these proteolytic needs can be met by the master recycling machinery called the 26S proteasome (Ciechanover and Kwon 2015; Mayer 2000; Meyer-Schwesinger 2019; Hnia et al. 2019; Coux et al. 1996; Finley and Prado 2019; Bard et al. 2018; Finley et al. 2016; Collins and Goldberg 2017; Voges et al. 1999; Tanaka 2009). The 2.5-MDa 26S proteasome counteracts the ribosome, by controlling the fate of synthesized proteins, and tightly regulates intracellular protein contents (Livneh et al. 2016; Collins and Goldberg 2017). To date, the 26S proteasome remains the largest, most enigmatic degradation machinery known and is ubiquitously found in all eukaryotic kingdoms (Darwin 2009; Muller and Weber-Ban 2019).
To be degraded by the proteasome holoenzyme, the targeted proteins must first be covalently modified by ubiquitin moieties via a cascade of three types of enzyme, namely, the E1 ubiquitin-activating enzyme, the E2 ubiquitin-conjugating enzyme, and the E3 ubiquitin ligase (Ciehanover et al. 1978; Ciechanover et al. 1980a, b; Hershko et al. 1979, 1980). Ubiquitin is a highly conserved protein with 76 amino acids ubiquitously found in eukaryotic cells (Wilkinson et al. 1980; Goldstein et al. 1975). Regulated modification of protein substrates with ubiquitin initiates their recognition and breakdown by the proteasome in an ATP-dependent fashion (Etlinger and Goldberg 1977; Arrigo et al. 1988; Wilkinson et al. 1980; Hershko et al. 1980). Mammalian cells contain only very few E1 enzymes, several E2 enzymes and at least several hundreds of different E3 ligases (Buetow and Huang 2016; Zheng and Shabek 2017; Hua and Vierstra 2011). The hierarchical cascade of ubiquitylation enzymes allows orthogonal regulation of hundreds of substrates in parallel. A large network of proteins involved in ubiquitylation and ubiquitin-mediated degradation by the 26S proteasome constitutes the ubiquitin-proteasome system (UPS) (Mayer 2000; Ciechanover 2005; Finley and Prado 2019; Livneh et al. 2016). Through the UPS, the proteasome elegantly regulates all major aspects of cellular processes, such as the cell cycle, gene expression, signal transduction, immune response, apoptosis and carcinogenesis (Ciechanover 2005). To serve as the sole terminal “processor” for myriad ubiquitylation pathways, the proteasome evolved exceptional adaptability in dynamically organizing ubiquitin receptors, shuttle factors, deubiquitinases, AAA-ATPase unfoldases, ubiquitin ligases and a large network of proteins to enable substrate selectivity, processing efficiency and precision of intracellular regulation (Finley and Prado 2019). The proteasome holoenzyme is one of the most dynamic enzymes known. The inner working of the proteasome is among the most sophisticated, enigmatic mechanisms of enzymatic machinery.
Indeed, owing to extremely complex dynamics in the 26S proteasome, the elucidation of its atomic structures took about four decades following its first discovery in the late 1970s (Etlinger and Goldberg 1977) despite continuous efforts by the worldwide research community and several generations of scientists (Bard et al. 2018). Building upon recent cryogenic electron microscopy (cryo-EM) studies on the human proteasome dynamics (Frank 2006; Chen et al. 2016a; Lu et al. 2017b; Zhu et al. 2018), an eventual breakthrough in visualizing substrate-processing dynamics of the human 26S proteasome at the atomic level (Dong et al. 2019) revealed the inner working of this sophisticated holoenzyme for the first time, clarified many long-term outstanding problems in the mechanisms by which a polyubiquitylated substrate is recognized, deubiquitylated, unfolded and degraded by the proteasome. These discoveries were partly echoed or complemented by separate structural studies of the substrate-bound yeast proteasome (de la Pena et al. 2018) and of other related systems in lower organisms (Puchades et al. 2017; Yu et al. 2018; Cooney et al. 2019; Majumder et al. 2019; Puchades et al. 2019; Twomey et al. 2019; Ripstein et al. 2020; Ding et al. 2019). These studies together establish the “central dogma” of the proteasome, thus revolutionizing our mechanistic understanding of the UPS. Comparisons among these studies further provide insights into conservation and variation of the operating principles used by proteolytic machineries in general. In this chapter, I offer a thorough account for our current understanding on the structure, dynamics and function of the 26S proteasome. Throughout, all descriptions are presented in the context of the human 26S proteasome in default, for brevity, unless otherwise explicitly stated.
Overview of the Ubiquitin-Proteasome System
Ubiquitylation
Ubiquitin and its polymerized form serve as the degradation signals for substrate recognition by the 26S proteasome (Komander and Rape 2012). Ubiquitin is an essential member in the protein family that shares a globular β-grasp fold of around 70 amino acids with a surface-exposed C-terminal glycine residue. Canonical ubiquitylation of a protein substrate is achieved via an isopeptide bond connecting the C-terminal glycine of ubiquitin with the ε-amino group of any lysine side chain exposed on the solvent-accessible surface of the substrate (Ciehanover et al. 1978, 1980a, b; Hershko et al. 1979, 1980). In rare non-canonical cases, covalent attachment of ubiquitin to cysteine, threonine, serine or the N-terminal amino group in the substrate have also been observed (Kravtsova-Ivantsiv and Ciechanover 2012).
Ubiquitylation involves sequential, cascaded actions of the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin ligase (E3), and the ubiquitin chain-elongation enzyme (E4) families (Fig. 1.1) (Koegl et al. 1999). The E1 enzyme catalyzes ATP-dependent activation of ubiquitin and isopeptide bond formation. The activated ubiquitin is then transferred to an E2 enzyme. The activated intermediate of E2-ubiquitin functions as the ubiquitin donor to the E3 ligase enzyme, which binds both the substrate and E2-ubiquitin intermediate via different structural motifs and recruits the ubiquitylation target as the substrate for the proteasome (Baek et al. 2020). Four families of E3 ligases have been discovered and classified into HECT, U-box, RING and RING-between-RING (RBR), based on their molecular architecture and enzymatic mechanisms. To enable selectivity and diversity in substrate processing, eukaryotic cells have evolved at least hundreds of distinct E3 ligases that feature a broad spectrum of substrate-interacting regions resembling a limited number of structural scaffolds (Buetow and Huang 2016; Zheng and Shabek 2017; Hua and Vierstra 2011). For example, the multi-subunit cullin-RING ligases (CRLs) in the RING family of E3 enzymes employ one of several cullin isoforms to assemble the complex (Baek et al. 2020). The remarkable diversity of the E3 enzymes allows the UPS to operate in various intracellular contexts for tightly regulating specific cellular activities via proteasomal degradation (Samant et al. 2018). In some cases, the first set of ubiquitin molecules attached to a protein substrate is further ubiquitylated so that the substrate becomes modified with longer polyubiquitin chains. The additional ubiquitin moieties can be attached by the same E3 or by a different ubiquitin ligase E4 (Koegl et al. 1999) that can only add more ubiquitin to substrate-conjugated monoubiquitin or polyubiquitin chains such as UBE3C/Hul5 and Ufd2 (Crosas et al. 2006; Hanzelmann et al. 2010).
Overview of the ubiquitin-proteasome system. Ubiquitylation is accomplished by the covalent conjugation of ubiquitin to lysine residues exposed on the surface of targeted substrates. Ubiquitin is first activated by the E1 in the presence of ATP. The ubiquitin is then transferred from E1 to E2. A ubiquitin ligase (E3) recruits the ubiquitin-bound E2 enzyme and a substrate to transfer the ubiquitin from E2 to the substrate. A specific type of ubiquitin ligase (E4) elongates the ubiquitin chains after monoubiquitin or polyubiquitin. The formation of Lys48-linked polyubiquitin chains, the main signal for proteasomal degradation, is mediated by successive cycles of ubiquitin conjugation. Monoubiquitylation could be sufficient for proteasome targeting in certain scenario. The monoubiquitin and polyubiquitin are removed from the substrate by the deubiquitinases (DUBs) associated with the proteasome. After being translocating into the proteasome, substrates are cleaved into short peptides, which are further broken down to amino acids by aminopeptidases (APPs). Released free polyubiquitin molecules are further recycled by cytosolic DUBs for another round of ubiquitylation
Many substrates of the proteasome are ubiquitylated on more than one lysine residue. The ubiquitylation site on a substrate itself is not tightly bound to the E3 so that structural flexibility of the ubiquitylation sites would allow several different lysine residues to interact with the ubiquitin bound to the E2 in the E2-E3-substrate intermediate complex (Baek et al. 2020). Through the conjugation cascade, the substrates could be modified with only one ubiquitin, with several ubiquitin molecules at distinct sites, or with polyubiquitin chains that are concatenated covalently via any surface-exposed lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63) or the N-terminal α-amino group of methionine (Met1) in ubiquitin. These three types of ubiquitin conjugation are referred to as monoubiquitylation, multiubiquitylation and polyubiquitylation, respectively (Kirisako et al. 2006; Xu et al. 2009; Yau et al. 2017). With the growth of the polyubiquitin chains, branched or tree-like heterotypic chain topologies have been observed (Yau et al. 2017). Such a complexity in ubiquitin polymerization generates highly diverse, heterogeneous degradation signals and allows ubiquitylation to regulate myriad cellular functions, some of which are not directly linked to proteolysis (Oh et al. 2018). The complexity of the ubiquitin signals is reversely “decoded” through the proteasome-dependent recruitment of several ubiquitin receptors (Husnjak and Dikic 2012; Lu et al. 2015; Oh et al. 2018), and many deubiquitylating enzymes (DUBs) capable of selectively processing substrates with various types of ubiquitin linkages (de Poot et al. 2017; Clague et al. 2019; Komander et al. 2009).
26S Proteasome Holoenzyme
As the sole proteolytic “central processor” in the UPS, the 26S proteasome holoenzyme assembles through noncovalent association of two subcomplexes, core particle (CP) and regulatory particle (RP). The RP caps either or both sides of the CP cylinder (Fig. 1.2). The CP, also known as the 20S proteasome, ubiquitously exists in prokaryotes and eukaryotes (Coux et al. 1996). The RP subcomplex, also known as PA700 or 19S, is a vital, indispensable proteasome activator found in all eukaryotic cells. The RP recruits, deubiquitylates, unfolds and translocates protein substrates into the CP chamber for degradation in an ATP-dependent fashion. In archaeal cells, one of the equivalent ATP-dependent activators is the proteasome-activating nucleotidase (PAN) complex, an ortholog of the eukaryotic proteasomal AAA-ATPase (Benaroudj et al. 2003; Smith et al. 2005). Several ATP-independent activators, typically PA28 (11S/REG) and PA200 (Blm10), can also bind the CP for proteasome activation (Stadtmueller and Hill 2011).
Overall architecture of the 26S proteasome. a The cryo-EM reconstruction of the doubly capped 26S proteasome. The CP is shown as cyan. The RP is marked by the dashed box. b Illustrative anatomy of the RP-CP subcomplex structure, with black silhouettes representing the central cross-section along the ATPase channel colored opaque blue. Dashed green curve illustrates the substrate-translocation pathway across the channel to the proteolytic sites in the CP chamber. Only upper half of the CP is shown
Core Particle
Proteasomal peptidase activities are housed in the CP. The CP is composed of 14 α-type and 14 β-type protein subunits (Lowe et al. 1995; Groll et al. 1997; Arrigo et al. 1988; Dong et al. 2019). In eukaryotic cells, there are seven distinct α-type or β-type subunits. Each type of subunit assembles into a heteroheptameric ring. Four such rings stack into a cylinder-shaped barrel in an α1–7β1–7β1–7α1–7 arrangement with an approximately two-fold symmetry (Lowe et al. 1995; Groll et al. 1997). In prokaryotic cells, there is only one gene for either the α-type or β-type subunit that assembles into a homoheptameric ring. Similar to the proteasomal CP, several other proteolytic proteins also assemble into a barrel-shaped proteolytic chamber, such as HslV (Bochtler et al. 1997) and ClpP (Wang et al. 1997). In eukaryotes, each β-ring houses three proteolytically active threonine residues in β1, β2 and β5 and forms the catalytic chamber. The active sites of the peptidase are sequestered inside the catalytic chamber. Heptameric α-rings, positioned on both sides of the catalytic chamber, control substrate entry into this space. Opening of an axial channel within the α-ring is allosterically controlled by the interactions of the CP with the 19S RP (PA700) or other activators such as PA200 and PA28 (Whitby et al. 2000; Zhu et al. 2018; Dong et al. 2019).
In lymphoid tissues or other interferon-γ (IFN-γ) stimulated cells, three constitutively expressed β-subunits are substituted with the subtype subunits β1i, β2i and β5i to assemble a variation of the CP named the immunoproteasome (Ferrington and Gregerson 2012). Different subtypes of β subunits may be mixed and combined in a single proteasome assembly to form an intermediate-type CP that exhibits different enzymatic properties (Dahlmann et al. 2000). Other CP subtypes such as thymus-specific (Murata et al. 2007) and testis-specific CPs (Uechi et al. 2014) have been observed to contain subunits β5t and α4s, which replace β5 and α4, respectively. These cell-type specific CPs were also termed the thymoproteasome (β5t) and spermatoproteasome (α4s), respectively (Murata et al. 2007, 2018). Subtype variations of the CP fine-tune its catalytic activity to meet tissue-specific functional needs.
Regulatory Particle
The RP consists of at least 18 protein subunits and regulates substrate recognition, recruitment, and unfolding in an ATP-dependent manner. The RP is structurally divided into two subcomplexes named the lid and the base that may assemble separately (Glickman et al. 1998). The lid subcomplex comprises 9 Regulatory Particle Non-ATPase (RPN) subunits, i.e., RPN3 (PSMD3/S3), RPN5 (PSMD12), RPN6 (PSMD11/S9), RPN7 (PSMD6/S10), RPN8 (PSMD7/S12), RPN9 (PSMD13/S11), RPN11 (PSMD14/Poh1/Pad1), RPN12 (PSMD8/S14), and RPN15 (PSMD9/Dss1/Sem1) (Table 1.1). Six subunits (RPN3, RPN5, RPN6, RPN7, RPN9 and RPN12) exhibits similar architecture featuring an N-terminal Proteasome-CSN-Initiation factor 3 (PCI) domain (Hofmann and Bucher 1998). Two subunits (RPN8 and RPN11) feature an MPR1-PAD1 N-terminal (MPN) domain and form an MPN dimer (Tran et al. 2003). All of the eight subunits are decorated with a C-terminal α-helical domain separated via a structured linker from their core PCI/MPN domains. The lid subcomplex is organized into a horseshoe-like architecture through an elaborate bundle that is composed of the C-terminal α-helical domain of each lid subunit except RPN15 (Estrin et al. 2013).
The base subcomplex includes RPN1 (PSMD2/S2), RPN2 (PSMD1/S1), and six paralogous, distinct Regulatory Particle ATPase (RPT) subunits—RPT1 (PSMC2/S7), RPT2 (PSMC1/S), RPT3 (PSMC4/S6), RPT4 (PSMC6/S10), RPT5 (PSMC3/S6a) and RPT6 (PSMC5/S8)—from the classic ATPases Associated with diverse cellular Activities (AAA) family (Table 1.1). The six RPT subunits of the base share a general domain organization, consisting of an N-terminal coiled-coil (CC) domain, an oligonucleotide- and oligosaccharide-binding (OB) domain, and a C-terminal AAA domain. They form a heterohexameric ATPase ring that acts as a mechanical motor. The well-folded domains of a substrate are unfolded by the mechanical force produced by the ATPase motor ring and translocated via the central pore of the ATPase ring by harvesting the chemical energy of ATP hydrolysis catalyzed by the ATPases. Thus, the ring-like RPT hexamer is also referred to as an unfoldase or translocase in the literature (Bard et al. 2018).
The first step of substrate processing by the proteasome is recognition of a ubiquitylated substrate, which is mediated by the ubiquitin receptors within the base, including RPN1 (PSMC2/S7) (Shi et al. 2016), RPN10 (PSMD4/S5a) (Deveraux et al. 1994; van Nocker et al. 1996), and RPN13 (ADRM1) (Husnjak et al. 2008; Schreiner et al. 2008). RPN13 can reversibly associate with RPN2 and is sub-stoichiometric in the endogenously purified human proteasome. In addition to the ubiquitin receptors intrinsically residing in the proteasome, ubiquitylated substrates can also be recruited and delivered to the proteasome by extrinsic ubiquitin receptors consisting of ubiquitin-like (UBL) and ubiquitin-associated (UBA) domains, including RAD23, DSK2, and DDI1 (Elsasser and Finley 2005; Elsasser et al. 2004; Zhang et al. 2009a). These UBL-UBA proteins interact with the intrinsic ubiquitin-binding sites in the proteasome via their UBL domain, functioning as shuttle factors that diversify the routes of decoding the ubiquitin signals for substrate selection and recognition.
During substrate engagement, an unstructured initiation region of the substrate is recognized by the pore loops of the RPT subunits (Prakash et al. 2004; Yu and Matouschek 2017; Bard et al. 2019; Dong et al. 2019). To allow substrate translocation into the CP, conjugated ubiquitin chains are cleaved from substrates by either the DUB RPN11 (Verma et al. 2002; Yao and Cohen 2002; Worden et al. 2017; Dong et al. 2019) or other auxiliary DUBs like USP14 (ubiquitin-specific protease 14) (Lee et al. 2016) and UCH-L5 (ubiquitin carboxyl-terminal hydrolase isozyme L5) (Vander Linden et al. 2015). RPN11 is a zinc-dependent DUB in the JAB1/MPN/MOV34 (JAMM) family protein (Ambroggio et al. 2004) and is intrinsically located above the OB ring, guarding the entry of substrates into the ATPase ring and CP. It catalyzes en bloc substrate deubiquitylation when a substrate is threaded into the ATPase ring.
The axial entrance of the CP proteolytic chamber, named the CP gate, is closed in the resting state (Chen et al. 2016a; Huang et al. 2016; Schweitzer et al. 2016). Opening of the CP gate is allosterically triggered by insertion of the C-termini of all RPT subunits except RPT4 into the outward pockets of the α-ring (α-pockets) located between adjacent α-subunits (Chen et al. 2016a; Zhu et al. 2018; Dong et al. 2019; Eisele et al. 2018; de la Pena et al. 2018; Smith et al. 2007; Rabl et al. 2008). The PA200 and PA28 complexes are two alternative CP-activating regulators that can replace the 19S RP in an assembled holoenzyme (Table 1.1). They trigger CP gate opening in an ATP-independent manner due to lack of ATPase domain. PA200 is a monomeric protein of ~250 kDa and conserved from the yeast to human (Schmidt et al. 2005; Blickwedehl et al. 2008). The PA28 family is expressed in higher eukaryotes and some unicellular eukaryotes such as trypanosomes. In mammalian cells, three isoforms (PA28α, PA28β, PA28γ) form two kinds of activators with distinct properties of expression, localization and activation. PA28α and PA28β are 28-kDa proteins that assemble into heteroheptameric ring structures in vertebrates (Ma et al. 1992; Dubiel et al. 1992) (Table 1.1).
History of Proteasome Structure Determination
X-Ray Crystallography of Proteasome Components
It has been half century since the first image of the human CP was recorded by negative-stain electron microscopy when its function was unknown (Fig. 1.3) (Harris 1968). The gradual elucidation of the proteasome structure and function was propelled and accompanied by innovations in a broad set of methods and tools in biochemistry and structural biology. Because the proteasomal CP is highly stable overall, it became the first key component to be crystallized. In the 1990s, both atomic structures of archaeal and yeast 20S proteasome were solved by X-ray crystallography (Lowe et al. 1995; Groll et al. 1997). The crystal structure of the human CP was solved much later (Harshbarger et al. 2015; Schrader et al. 2016). Crystal structures of the isolated CP or immunoproteasome CP are available for several species (Groll et al. 1997; Unno et al. 2002; Harshbarger et al. 2015; Huber et al. 2012). These structural works defined three conserved proteolytic sites in the β1, β2 and β5 subunits, laying an important foundation for understanding the activation of the CP and the mechanism of proteolysis. However, all these high-resolution CP structures represent a basal resting state with a closed gate in their α-rings. The first yeast CP with an open gate in the α-ring was determined by X-ray crystallography in the presence of activation by the PA26 (11S) regulator (Whitby et al. 2000), whereas the activated human CP in an open-gate state was only determined much later by cryo-EM (Chen et al. 2016a; Zhu et al. 2018; Dong et al. 2019).
Timeline and milestones in the elucidation of structure and function of the 26S proteasome. In 1968, the first image of the human proteasome CP was captured by negative-stain EM when there was no knowledge about its function (Harris 1968). In the 1970s, concerted discoveries revealed the functional role of the proteasome by studying ATP- and ubiquitin-dependent degradation system in eukaryotic cells (Etlinger and Goldberg 1977; Ciehanover et al. 1978; Hershko et al. 1979; Ciechanover et al. 1980a, b; Hershko et al. 1980; Wilkinson et al. 1980). In the 1980s, the approaches of purification of mammalian 26S proteasome were developed, paving the way to its structural analysis (Tanaka et al. 1983; Hough et al. 1986, 1987). In 1993, the first image of the mammalian 26S proteasome was captured by negative-stain EM (Peters et al. 1993). Soon after, X-ray crystallography revealed the proteasome CP structures in both Archaea and yeast (Lowe et al. 1995; Groll et al. 1997). In 2010, the first subnanometer resolution structure of a yeast 26S proteasome was reconstructed by cryo-EM (Bohn et al. 2010). During 2015–2017, the first high-resolution structures of the substrate-free human proteasome were determined by cryo-EM, which revealed six coexisting conformations (Chen et al. 2016a; Huang et al. 2016; Schweitzer et al. 2016; Zhu et al. 2018), whereas cryo-EM structures of their yeast counterparts were also determined at the similar resolution range in the following years (Eisele et al. 2018; Wehmer et al. 2017; Ding et al. 2017, 2019). In 2018, the first atomic structures of the substrate-engaged human 26S proteasome in seven functional states were determined to higher resolution by cryo-EM (Dong et al. 2019), whereas the yeast counterparts in four substrate-engaged states were analyzed at slightly lower resolutions (de la Pena et al. 2018)
In stark contrast to conformational stability of the CP, the structure of the RP is highly dynamic and samples an extensively expanded conformational landscape (Lu et al. 2017b; Unverdorben et al. 2014; Chen et al. 2016a; de la Pena et al. 2018; Zhu et al. 2018; Dong et al. 2019). Thus, the RP structure in the 26S proteasome has completely evaded X-ray crystallography. However, many but not all of the RP subunits were separately solved in certain domains or regions by X-ray crystallography or nuclear magnetic resonance (NMR) (He et al. 2012; Pathare et al. 2012; Boehringer et al. 2012; Riedinger et al. 2010; Worden et al. 2014; Pathare et al. 2014; Zhang et al. 2009b, c; Schreiner et al. 2008; Shi et al. 2016). Despite its simplicity, the archaeal PAN complex, an ATPase homohexamer, is thought to represent an evolutionary precursor to the eukaryotic proteasomal ATPases. The OB and AAA domains of the PAN ATPase have been separately crystallized and solved in a fragmented form (Zhang et al. 2009b, c).
Early Electron Microscopy Studies of Proteasome at Low Resolution
The 19S RP in the 26S proteasome is highly dynamic and prevents the 26S proteasome from being crystallized. Thus, electron microscopy reconstruction became the only option to obtain the molecular shape of the 26S proteasome in the 1990s (Peters et al. 1993; Frank 2006). The first cryo-EM reconstruction of the 26S proteasome from Drosophila at 1–2 nm resolution was documented in 2009 (Nickell et al. 2009). Following the improvement of single-particle reconstruction method, the cryo-EM structure of substrate-free Schizosaccharomyces pombe 26S proteasome at a subnanometer resolution (9.1 Å) was reported (Bohn et al. 2010). This level of resolution was insufficient to allow direct identification of subunit organization. Thus, other methods in biochemistry and cross-linking mass spectrometry were used in an integrative fashion to define the subunit architecture of the RP (Beck et al. 2012; Lander et al. 2012; Lasker et al. 2012; da Fonseca et al. 2012). Using the crystal structures of individual RP subunits, the first pseudo-atomic model for the yeast 26S proteasome was built using ~ 8 Å maps (Beck et al. 2012). In the following years, additional cryo-EM reconstructions of the yeast proteasome at subnanometer resolutions were analyzed under various substrate-free conditions and revealed three distinct conformational states (designated s1, s2 and s3), which were hypothetically interpreted as the substrate-accepting, commitment and translocating states, respectively (Matyskiela et al. 2013; Unverdorben et al. 2014; Sledz et al. 2013). Meanwhile, a 9-Å map of the substrate-bound yeast proteasome was reconstructed but the density of substrate inside the proteasome was averaged out (Matyskiela et al. 2013). Nevertheless, the overall conformation of the substrate-bound yeast proteasome resembles that of state s3, although they are not completely identical. The limited resolution, however, precludes understanding of the structural mechanisms for ATP-dependent degradation by the 26S proteasome (Forster et al. 2013). Inter-subcomplex regulation, coordinated ATP hydrolysis in the AAA-ATPases, and substrate-proteasome interactions remained particularly elusive in these studies.
High-Resolution Cryo-EM Studies of Substrate-Free Proteasome
My colleagues and I started working on the human proteasome structure in 2014. Our first near-atomic resolution reconstruction of proteasome was determined in the late 2015 (Chen et al. 2016a). However, it showed closed gates on both sides of the CP, in contradiction to early conclusions derived from the lower resolution cryo-EM maps (Beck et al. 2012; Lander et al. 2012; Lasker et al. 2012; da Fonseca et al. 2012). To clarify this issue, additional analysis was conducted to test the hypothesis on a possible coexisting conformational state with an open CP gate. A focused classification strategy was devised by subtracting the CP density from the original raw single-particle cryo-EM images, following which exhaustive 3D classification eventually sorted out four coexisting conformations (designated states SA, SB, SC and SD) of the human proteasome in presence of ATP/Mg2+, among which only one state SD was open in the gate of CP (Chen et al. 2016a). This allowed us to observe that insertion of five RPT C-terminal tails into the CP albeit at a moderate resolution (6–8 Å) for the first time (Chen et al. 2016a). The cryo-EM structure of the human proteasome in state SA but no other states were also reported at comparable resolutions by two other groups (Huang et al. 2016; Schweitzer et al. 2016). In the following year, a high-resolution structure of the yeast proteasome in state s1 (analogous to human SA) was reported, along with an open CP state s4 (analogous to human SD) at ~8-Å resolution discovered when the yeast proteasome was bound to ATP analogs instead of ATP (Wehmer et al. 2017).
Following the first reconstruction of human proteasome at the atomic level, several groups set out to determine the atomic structures of the substrate-bound proteasome holoenzyme. However, the exceptional dynamics prevent the complex from being reconstructed at high resolution by using the same cryo-EM procedure practiced in the determination of the substrate-free proteasome in the resting state (SA). It was soon realized that unprecedented challenges in cryo-EM and biochemical methodology must be conquered before one can obtain an atomic structure of the substrate-bound proteasome. First, how can one capture the complex before substrate degradation is completed? Second, how can one deconvolute the exceptional conformational heterogeneity in the sample not otherwise commonly encountered? Third, how can one push as many proteasome conformations as possible to the near-atomic resolution range (2.5–3.6 Å)? To confront these formidable challenges and to find a feasible path forward, we devised two pilot studies along with several parallel cryo-EM methodology developments (Xu et al. 2016; Zhu et al. 2017; Wu et al. 2017; Wang et al. 2019), with the anticipation of gaining new ideas for devising novel methods for solving the substrate-bound proteasome at the atomic level.
In one study, the free RP complex was analyzed by cryo-EM in states presumably prior to its assembly with the CP. In the free RP, the AAA-ATPase ring exhibited continuous motion of a broad conformational landscape, captured in seven conformational states at ~9 Å resolution in the AAA-ATPase but 4.6 Å for the rest of the free RP (Lu et al. 2017b). In another study, the 26S proteasome was analyzed in the presence of ATPγS that replaces ATP. It was aimed to improve the open-gate state to high resolution (Zhu et al. 2018). Surprisingly, ATPγS induced a dramatic shift in the conformational landscape of the proteasome from state SA to SD. To improve the resolution of state SD, we had to further classify the dataset corresponding to state SD into three distinct sub-states SD1, SD2 and SD3, which were then improved to 4.3–4.9 Å (Zhu et al. 2018). Averaging of the three sub-states instead gave rise to a 6-Å resolution with smeared features in the center of the ATPase ring. These studies taught us an important lesson that 3D classification of a highly heterogeneous dataset with significantly improved clustering accuracy is crucial for achieving higher resolution for those lowly populated conformational states. The observations of six coexisting states of the ATPγS-bound human proteasome was soon echoed by the discovery of states s5 and s6 of the yeast proteasome (Eisele et al. 2018), confirming the conservation of the proteasome dynamics in the absence of a substrate.
Visualizing Atomic-Level Dynamics of Functional Proteasome
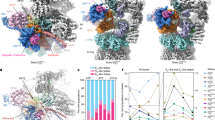
In the early summer of 2018, seven distinct states of the substrate-engaged proteasome at 2.8–3.6 Å resolution (Fig. 1.4), which are referred to as EA1, EA2, EB, EC1, EC2, ED1 and ED2, eventually emerged after a few years of marathon-like repeated protein purification, data collection and analysis (Dong et al. 2019), in sharp contrast to some of our other cryo-EM structures solved in a month (Zhang et al. 2015). These exciting results would not be made possible without several lines of methodology innovation and improvement. First, instead of completely replacing ATP with ATPγS or nucleotide analogs like several studies (Sledz et al. 2013; Wehmer et al. 2017; Eisele et al. 2018; Zhu et al. 2018; Yu et al. 2018), a novel “nucleotide-substitution” strategy that dilutes ATP with ATPγS in a time-dependent manner was deliberately devised to capture the functional proteasome conformations in action that are as diverse as possible (Dong et al. 2019). The proteasome was first primed with a bona fide polyubiquitylated substrate Sic1PY and ATP/Mg2+ for 30 s, then exchanged in a buffer containing both ATP and ATPγS at a 1:1 stoichiometric ratio (Dong et al. 2019). The choice of 30 s delay in ATPγS dilution after substrate-proteasome mixing was based on the approximate half-life of the substrate degradation reaction, which is consistent with a recent kinetic measurement on proteasomal degradation (Bard et al. 2019). This strategy decelerated the hydrolysis activity of AAA-ATPases by partially replacing ATP with ATPγS in the proteasome. This is expected to maximally preserve the conformational landscape and native heterogeneity of the substrate-engaged proteasome being captured before the completion of substrate degradation reactions (Dong et al. 2019) (Fig. 1.4).
Seven cryo-EM maps of the substrate-engaged Homo sapiens 26S proteasome at 2.8–3.6 Å resolution capture the key intermediate steps of substrate processing and provide important insights into the chemical cycle of proteasome-mediated degradation (Dong et al. 2019)
Second, to overcome the difficulties of cryo-EM data analysis entailed by an extreme degree of conformational heterogeneity in this case, we decided to collect a considerably larger cryo-EM dataset (44,664 raw micrographs) that is necessary for improving resolutions for those lowly populated conformational states (Dong et al. 2019), which would otherwise be either invisible or of low resolution with a normally sized dataset. Note that a much larger dataset would drastically complicate the computational task of 3D classification rather than straightforwardly ensuring higher classification accuracy or higher resolution, because it could readily break the upper limit of high-performance computational resources, causing frequent failure of popular cryo-EM software. In fact, more data can effectively reduce resolution rather than increasing it if the data classification accuracy is not warranted (Wu et al. 2017).
Third, we extensively used the latest data processing tools developed in my laboratory, including deep-learning-based particle selection (Zhu et al. 2017), manifold-learning-based 2D clustering (Wu et al. 2017), and hardware-accelerated 3D classification to optimize the data processing. Without these new computational tools, it could have taken at least another year or longer to analyze the same dataset. Furthermore, building upon lessons learnt from previous studies, we further fine-tuned a hierarchically focused 3D classification strategy with stepwise zoomed-in masking and eventually sorted out seven conformations of the substrate-bound proteasome covering the nearly complete cycle of substrate processing from substrate recruitment and deubiquitylation to processive degradation (Dong et al. 2019).
Before fully committed to the “nucleotide substitution” strategy, there was a debate in my group regarding whether we should instead follow a more conventional, lower-risk strategy of stabilizing the substrate-bound proteasome by inhibiting the deubiquitylating activity of RPN11, which was previously used in a cryo-EM study of the substrate-bound yeast proteasome (Matyskiela et al. 2013). But concerns were that this treatment might be insufficient to lock the proteasome into a single conformation, could fail in capturing the proteasome in functional steps other than substrate translocation and also potentially create some artificial structural features that are not physiologically relevant. These concerns turned out to be mostly the case when four alternative cryo-EM structures of the substrate-bound yeast proteasome at 4.2–4.7 Å resolution were obtained by inactivating the yeast Rpn11 with the inhibitor ortho-phenanthroline (de la Pena et al. 2018). The inhibition of the yeast Rpn11 results in certain structural features in state 1D* that are potentially off-pathway. However, the AAA-ATPase structures in states 5D, 5T and 4D of the substrate-engaged yeast proteasome are remarkably comparable to their human counterparts in states ED1 and ED2 within the limit of their reconstruction quality and resolution (de la Pena et al. 2018; Dong et al. 2019). This suggests that the molecular mechanisms of substrate processing by the proteasome must be highly conserved throughout the eukaryotic kingdom.
With hindsight, the proteasome has served as an extraordinary model system or “gold-standard trial” for furthering methodology development in cryo-EM for its potential in solving atomic-level dynamics of complex enzymes (Dong et al. 2019). By retrospective comparison, it appears that the structural determination of functional substrate-engaged proteasome was perhaps much more difficult than those of the ribosome and spliceosome, the other two well-known dynamic protein complexes. The conventional cryo-EM methodology presumes a homogeneous core structure or few dominating conformations that can afford high-accuracy image alignment leading to high-resolution reconstructions. However, the substrate-engaged proteasome samples a significantly expanded, highly frustrated energy landscape and severely violates the presumption of the conventional cryo-EM methodology. Misclassification of conformers was found to be the major cause for limited resolutions when the proteasome samples an expanded energy landscape (Zhu et al. 2018). However, improved curation in initial particle ensemble with deep learning, higher accuracy in data clustering using manifold learning, a fine-tuned, object-optimized 3D classification strategy and a significantly larger dataset all seem to contribute to the improvement of 3D classification accuracy beyond the realm of the conventional cryo-EM methodology. A more detailed account for the methodology advancement leading to the breakthrough of atomic structure determination of functional human proteasome will be provided elsewhere.
Structures and Dynamics of the Proteasome
Molecular Architecture of the Proteasome
Due to the highly dynamic nature of the proteasome, no single reconstructions of the 26S proteasome provided the best resolved local features for all components (Chen et al. 2016a; Huang et al. 2016; Schweitzer et al. 2016; Zhu et al. 2018; Dong et al. 2019; de la Pena et al. 2018; Eisele et al. 2018; Ding et al. 2019). Overall, the CP and AAA-ATPase motor in the RP were best resolved in state EA at 2.8-Å resolution; the non-ATPase subunits in the RP were best resolved in states ED1 and ED2 at 3.3 and 3.2 Å (Dong et al. 2019). The early high-resolution structures of the proteasome in state SA all presented poorer local density quality in the lid and the non-ATPase subunits in the base, where the local resolution did not go beyond the range of 4–8 Å (Chen et al. 2016a; Huang et al. 2016; Schweitzer et al. 2016; Zhu et al. 2018). This issue was addressed in the recent study at higher resolution in these subunits (Dong et al. 2019). Putting together, these atomic structures provide thus-far the nearly complete understanding of the 26S proteasome assembly. In this subsection, I intended to summarize the overall organization and architecture of the proteasome holoenzyme based on the highest resolution structures available. More details about conformational dynamics of the proteasome associated with distinct functional steps are discussed in the following subsections.
The Lid Subcomplex
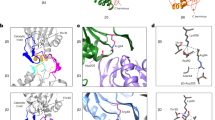
The ~370-kDa lid subcomplex in the RP has an overall dimension of 170 × 150 × 140 Å3. The architecture of the ~370-kDa lid subcomplex in the RP bears topological similarity to those of the COP9 signalosome (Lingaraju et al. 2014) and the eukaryotic translation initiation factor 3 (eIF3) (des Georges et al. 2015). The PCI domain of six RPN subunits (RPN9, RPN5, RPN6, RPN7, RPN3, and RPN12 from the left to right side in Fig. 1.5a) features N-terminal helical repeats (HR) connected to a winged-helix (WH) subdomain. The lid subcomplex is stabilized by two organizational centers (Fig. 1.5): a half-sized open ring formed through side-by-side oligomerization of three-stranded β-sheets of the WH subdomains from the six PCI proteins (PCI ring) (Fig. 1.5a), and an elaborate α-helical bundle comprising the C-terminal regions from all of eight PCI/MPN subunits (Fig. 1.5b). The N-terminal helical repeats of the six PCI proteins radially protrude from the PCI ring of 18-stranded β-sheet, forming a horseshoe-like architecture.
Atomic structures of the lid subcomplex in the human 26S proteasome (PDB ID: 6MSB) (Dong et al. 2019). a The lid-base interface lateral to the ATPase ring is composed of three local inter-subunit interfaces of RPN5-RPT3, RPN6-RPT6 and RPN7-RPT6, which are highlighted by dashed boxes 1, 2 and 3, respectively. b, c The atomic model of the lid subcomplex viewed from two perspectives orthogonal to that in (a). The central helical bundle is highlighted in cartoon representation. The rest of the lid structure is shown in a ribbon representation. The right inset zooms into the central helical bundle. The asterisk symbols label the N-terminal ends of the lid subunits
The PCI and MPN proteins form largely two separate subassemblies that are united in the central helical bundle. The central helical bundle is connected to the PCI ring from one side and to the MPN dimer of RPN8-RPN11 from the other side via structured linkers at the C-termini of the WH subdomains. Outside the helical bundle, the MPN dimer does not directly contact the PCI ring. It is separated by the base subunit RPN2 from one end and the von Willebrand factor type A (VWA) domain of another non-lid subunit RPN10 from the other end. Only the MPN domain of isopeptidase RPN11 in the lid subcomplex harbors a catalytic active zinc ion.
The Base Subcomplex
The base subcomplex is organized around the ATPase ring (RPT1-RPT6). The coiled-coil (CC) dimers of RPT1-RPT2, RPT3-RPT6, and RPT4-RPT5 harbor RPN1, RPN2 and RPN10, respectively. While the interaction between the RPT3-RPT6 CC domain and RPN2 was apparent in the cryo-EM maps of the proteasome in the resting state (SA), the other two pairs of interactions were insufficiently visualized due to the local conformational dynamics of RPN1 in the absence of a substrate (Chen et al. 2016a; Huang et al. 2016; Schweitzer et al. 2016; Zhu et al. 2018). The interface between RPT1-RPT2 CC and RPN1 was best visualized in the substrate-bound proteasome in states ED1 and ED2 at higher resolution (Dong et al. 2019).
RPN10 was originally thought to be part of the base (Glickman et al. 1998), but later considered to be neither part of the base nor the lid (Bard et al. 2018), based on the structural observation that the RPN10 VWA domain interacts directly with the lid subunits RPN8 and RPN9 in the proteasome and does not contact any base subunits in the resting state of the proteasome (Chen et al. 2016a). Nevertheless, cryo-EM structures of substrate-interacting proteasome did show a direct interaction of RPN10 with the base at the RPT4-RPT5 CC in state ED2 (Dong et al. 2019). Furthermore, the RPT4-RPT5 CC dimer seems to harbor auxiliary ubiquitin-binding sites that assist ubiquitin recognition by RPN10 and peptide-proximal ubiquitin transfer to RPN11 for deubiquitylation (Dong et al. 2019; Chen et al. 2020; Lam et al. 2002). The ubiquitin-interacting motifs (UIM) of RPN10 are structurally flexible and missing in all available cryo-EM reconstructions (Chen et al. 2016a; Huang et al. 2016; Schweitzer et al. 2016; Zhu et al. 2018; Dong et al. 2019). In the yeast proteasome, Rpn13 binds the C-terminal region of Rpn2 with a sub-stoichiometric ratio (Sakata et al. 2012).
The OB domains of six RPT subunits form a ring with an inner diameter of 10 Å (Fig. 1.6). This is the substrate entry port into the interior of the proteasome. Its narrow dimension indicates that the OB ring imposes the restriction against translocation of folded substrates. Each of the OB domains folds into a five-stranded β-barrel, whose axis lies orthogonal to that of the OB ring and is directed radially (Fig. 1.6h). Residues in loop L23 appear to impose the narrowest constriction in the central pore of the OB ring subcomplex. Despite being surrounded by highly dynamic structures, the OB ring mostly moves as a whole rigid body during ATP or substrate turnover.
The atomic model of the ATPase heterohexamer in the EA/SA state (PDB ID: 6MSB) (Dong et al. 2019). a The overall structure of the ATPase viewed from the perspective of the substrate entry port. The OB ring is in the foreground and is shown with transparent surface superimposed on a cartoon representation of the AAA domains. b Overall structure of the ATPase ring viewed from a perspective rotated 90° from that in panel (c). c–e The component structures of the OB ring (panel c), large AAA subdomains (panel d) and small AAA subdomains (panel e) viewed from the same perspective as that in panel (b). f–h The component structures of the OB ring (panel f), large AAA subdomains (panel g) and small AAA subdomains (panel h) viewed from the same perspective as that in panel (a)
The link between the CC and OB domains is marked by a conserved proline residue in the cis configuration in RPT2 (Pro106), RPT3 (Pro86) and RPT5 (Pro87) at homologous residue positions in the structure. Thus, a cis-Pro residue in every other subunit is essential for dimerization of the CC domains of three RPT heterodimers (RPT1-RPT2, RPT3-RPT6 and RPT4-RPT5). Alternating cis-Pro residues at this position have also been found in the archaeal PAN homohexamer—a remarkable instance of evolutionary conservation (Zhang et al. 2009b; Arcus 2002).
The AAA domains of RPT subunits have a common architecture, featuring an N-terminal α/β subdomain (large AAA subdomain) linked to a smaller C-terminal α-helical subdomain (small AAA subdomain) via a short loop. The concave pocket between the large and small AAA subdomain houses the nucleotide-binding site. In the resting state (SA) of the proteasome, the AAA domains of RPT subunits form a spiral staircase resembling a lock washer, in which RPT3 is located at the top position and RPT2 at the bottom, with RPT6 bridging RPT2 and RPT3 at the “split seam”. RPT6 possesses the most divergent fold in its AAA domain, consistent with its unique role in driving the initial substrate engagement (Dong et al. 2019).
The Lid-Base Interface
The interfaces between the lid and the base play crucial roles in coordinating substrate recognition, deubiquitylation and translocation (Greene et al. 2019). The lid-base interface is organized around the DUB RPN11, a metalloprotease fold, which is positioned at the substrate entrance of the OB ring of the ATPase heterohexamer. On one side of RPN8-RPN11, the MPN dimer is demarcated from RPN2, and the other side from the VWA domain of RPN10. There are three types of lid-base interfaces: (1) the central interface between RPN11 and the CC-OB domains of the ATPase ring; (2) the interface defined by the CC domains of the adjacent RPT3 and RPT6 subunits, which project distally from the central interface; and (3) the lateral interface between the AAA domains of ATPases and RPN5-RPN7. The interfaces between the lid and the ATPase ring bury a ~3900 Å2 surface area in total. Specifically, the CC-OB domains of RPT3-RPT6 contribute a ~3100 Å2 interfacial area with the lid. The RPT3-RPT6 CC domain is encircled by the helices in RPN2, RPN3, RPN8 and RPN11. This interfacial architecture appears to stabilize the lid-base association.
The lateral lid-base interface implicates distinct roles of interacting subunits in the conformational transitions that follow substrate engagement. The AAA domains of RPT3 and RPT6 bury ~1800 Å2 of interface area with RPN5-RPN7. One side of N-terminal helical repeats in the RPN7 PCI domain, consisting of four helix-connecting loops, makes extensive interaction (~940 Å2) with the RPT6 AAA domain. In contrast, the AAA domains of RPT3-RPT6 contact RPN5 and RPN6, with smaller buried footprints of ~340 Å2 and ~480 Å2, respectively. The interfaces of RPN7-RPT6, RPN6-RPT6 and RPN5-RPT3 may serve as “switches” to allosterically regulate the conformations of the RPT subunits, thus communicating between adjacent subcomplexes during substrate degradation (Greene et al. 2019; Dong et al. 2019).
The CP Subcomplex
The CP has a cylindrical shape, with overall dimensions of 148 × 113 × 75 Å3 (Fig. 1.7). While the archaeal CP is composed of 28 subunits arranged in four homoheptameric rings α7β7β7α7 coaxially stacked with D7 symmetry, the eukaryotic CP is assembled from four heteroheptameric rings, α1–7β1–7β1–7α1–7, with only C2 symmetry. The CP conceals the proteolytic active sites in the inner surface of β-ring. As defined by the characteristics of the P1 cleavage sites of chromogenic reporter groups, the Thr1 residues in β1, β2, and β5 subunits have been ascribed with caspase-like (acidic; peptidylglutamyl-peptide hydrolytic (PGPH)), trypsin-like (basic), and chymotrypsin-like (hydrophobic) activities. However, these specificities are not reflected in the cleavage-site pattern of its substrates that are cleaved at almost arbitrary positions.
The atomic structure of the human CP (PDB ID: 6MSB) (Dong et al. 2019). a The overall structure of the CP from a lateral view. The color codes of the subunits are displayed on the right. b The CP structure from a top view. c and d The atomic models of α1-subunit (c) and β1-subunit (d) are shown in cartoon representations, which are typically homologous among all the α- and β-type subunits. The names of the secondary structures are labelled
In eukaryotic cells, five β-subunits are synthesized as proproteins, which are proteolytically cleaved to become the mature forms during the final steps of proteasome assembly (Chen and Hochstrasser 1995; Seemuller et al. 1995a; Frentzel et al. 1994). The proforms of β1, β2, and β5 subunits are cleaved between Gly(-1) and Thr1, which liberates the active site at residue Thr1. β6 is cleaved between His(-10) and Gln(-9) and β7 is cleaved between Asn(-9) and Thr(-8). β1 and β3 are not further processed and remain in their primary translational forms. All seven α-subunits and seven β-subunits share the common α/β/β/α sandwich-like fold characteristic of Ntn (N-terminal nucleophile)-hydrolase superfamily (Oinonen and Rouvinen 2000). It exhibits two five-stranded antiparallel β-sheets sandwiched by three helices (H3, H4 and H5) on the top and two helices (H1 and H2) on the bottom. Structural differences between the subunits are entailed by the length variations (often by one or two residues) in the N-terminal and C-terminal regions, in turning loops and in long insertions bridging adjacent secondary structural elements. For instance, the subunits α3 and α4 have longer C-terminal helices (H5) sticking out from the CP surface toward the RP, potentially making direct contact with the base subunits.
The intersubunit interactions mediating the assembly of the CP can be categorized into α-cis, β-cis, α-trans-β, and β-trans-β contacts. The α-cis contacts are mediated by the N-terminal loop, the helix H0, the strand S7, the loop L, and the loop linking H2 with S5. The β-cis contacts are mediated by loop L, the N-terminus of helix H1, strand S7 and the turn connecting helix H3 and strand S8. The α-trans-β contacts are mediated by the helix-loop-helix motifs spanning H1 and H2, which interdigitate with the same motifs of two adjacent α-subunits. This mode of intersubunit contacts is conserved in both prokaryotes and eukaryotes (Lowe et al. 1995; Groll et al. 1997; Dong et al. 2019). β-trans-β contacts are more specifically formed by the C-terminal arm of β7 intercalating between β1′ and β2′ from adjacent β′-ring, and by the intercalation of β5 C-terminal arm with β3′ and β4′ in a similar configuration.
The CP gate refers to the central axial pore controlled by the N-terminal loops of α-type subunits (Groll et al. 2000). The CP stays in an inactive state in its free 20S form and in the resting state after its assembly with the RP. In this inactive state, the CP gate is blocked by stacking of N-terminal loops of only three α-type subunits, α2, α3 and α4. Particularly, the N-terminal loop of α3 occupies the majority of the space at the CP gate, resembling a linchpin that is stabilized by the N-terminal loops of α2 and α4. Opening of this gate is allosterically controlled by interactions between the CP and RP.
The RP-CP Interface
The RP associates with the CP through a symmetry-mismatched interface (~3600 Å2) between the hexameric AAA-ATPase motor and the heptameric α-ring in the resting state (SA). All RPT subunits but RPT6 directly contact the α-ring. The large void between the RPT6 and α2 subunits is sided by the N-terminal helical repeats in the PCI domain of RPN6. The helical repeats of RPN6 PCI domain protrudes down to the lateral surface of α2, making ~620-Å2 contact. Similarly, the N-terminal helical repeats of RPN5 PCI domain also make a lateral contact with the α1 subunit via a small footprint area (~50 Å2) (Chen et al. 2016a; Eisele et al. 2018; Zhu et al. 2018; Dong et al. 2019).
In the resting state, the conserved hydrophobic-Tyr-X (HbYX) motifs of RPT3 and RPT5 are docked into the α1-α2 and α5-α6 pockets, respectively (Chen et al. 2016a; Huang et al. 2016; Schweitzer et al. 2016) (Fig. 1.8). The terminal oxygen of the penultimate tyrosine in the HbYX motifs (Tyr417 in RPT3 and Tyr438 in RPT5) forms a hydrogen bond with the carbonyl oxygen in the mainchain of the glycine residue in the corresponding α-pocket (Gly20 in α1 and Gly19 in α5). Similar contacts have also been observed between PA26 and archaeal 20S proteasome precursors (Stadtmueller et al. 2010). These structural features explain the notion that RPT3 and RPT5 are the only two ATPase subunits whose C-terminal HbYX motifs are indispensable for assembly of the human 26S proteasome (Kim and DeMartino 2011). By contrast, in the yeast proteasome, the HbYX motifs of RPT2, RPT3 and RPT5 are inserted into the α-pockets (Tian et al. 2011; Stadtmueller et al. 2010; Ding et al. 2017; Wehmer et al. 2017).
Asymmetric opening of the CP gates in the 26S proteasome (Chen et al. 2016a; Zhu et al. 2018). a The cryo-EM density map of the CP in the SD state viewed from the side (left), the top (upper right), and the bottom (lower right). The RP density is not shown. The upper half of the CP faces the RP in the SD state and is in the open conformation. b The atomic models of the α-rings in the open (upper left) and closed (lower left) states in cartoon representation, viewed from the perspective of the RP-CP interface. The two panels in the right show the central parts of the α-rings in the open (upper right) and closed states (lower right). c Cryo-EM density map of the RP-CP interface in the open gate state, in which the local densities of the C-terminal tails of RPT1, RPT2, RPT6, RPT3 and RPT5 are highlighted by different colors for clarity. Two empty α-pockets are marked by asterisks. d The atomic models of α-rings in the closed (blue) and open (closed) states are superimposed
Complex Dynamics of the Proteasome
Common Traits of Proteasome Dynamics and Plasticity
The 26S proteasome was consistently observed to sample multiple coexisting conformers in both Homo sapiens and Saccharomyces cerevisiae (Unverdorben et al. 2014; Chen et al. 2016a; Luan et al. 2016; Wehmer et al. 2017; de la Pena et al. 2018; Eisele et al. 2018; Zhu et al. 2018; Ding et al. 2019; Dong et al. 2019). Although several conformers of the substrate-free proteasome preserve architectural features of the substrate-bound proteasome, certain conformers and crucial substrate interactions are not accessible in the absence of substrates. This underscores the importance of understanding the conformational landscape of the substrate-engaged proteasome under physiological conditions, which is characterized at large by the seven atomic structures of substrate-bound proteasome as summarized in Table 1.2. Because the conformational state in the RP and its interacting α-ring on one side of a doubly capped proteasome can be different from that of the interacting pair on the other side, the states listed in Table 1.2 refer to the conformations of either the RP-CP subcomplex or the singly capped proteasome. There is currently insufficient evidence showing functional difference between the RP-CP subcomplex in a doubly capped proteasome and the singly capped proteasome (Asano et al. 2015).
There are several common patterns of pronounced conformational changes occurring in the proteasome under distinct biochemical conditions. First, the CP conformation assumes states of either closed or open gate. The human CP structures with a closed gate are virtually identical in all corresponding states including EA1, EA2, EB, EC1, EC2, SA, SB and SC (Dong et al. 2019). The same is true for the human CP with an open gate in the remaining states. This reflects a bistability of the CP gate that has been also observed for the yeast CP. Second, the lid rotation is generally accompanied with the conformational transitions between different states. Most notably, a prominent lid rotation of 25–40° is commonly observed during state transitions of EB-to-EC (substrate-engaged) (Dong et al. 2019), SA-to-SB transition (substrate-free) (Chen et al. 2016a; Zhu et al. 2018), C1-to-C3 (tetraubiquitin-bound yeast proteasome) (Ding et al. 2019), and s1-to-s2 (substrate-free yeast proteasome) (Wehmer et al. 2017; Eisele et al. 2018; Ding et al. 2017). Third, the quaternary structural relationship between RPN11 and the OB ring is also approximately bistable. The RPN11 blocks the entrance of the OB ring in states EC1, EC2, ED1, ED2, SB, SC, SD1, SD2, SD3, 5D, 5T, 4D, C3-a, C3-b, s2, s3, s4, s5, and s6 (Dong et al. 2019; de la Pena et al. 2018; Eisele et al. 2018; Ding et al. 2019; Chen et al. 2016a; Zhu et al. 2018). By contrast, RPN11 is rotated to the side of the OB ring and leaves a wide entrance to the ATPase channel in all remaining states. Fourth, whenever the CP gate is open in states ED1, ED2, SD1, SD2, SD3, 5T, 4D, C3-a, C3-b, s4, s5, and s6, the conformations of the ATPase ring in these states are always remodeled and translated above the CP to positions that confer a better axial alignment of the ATPase channel with the CP gate as compared to the resting state (SA). Fifth, the pore-loop staircase is similarly observed in states ED1, 5D and 5T, whereas a different pore-loop staircase is comparably observed in states ED2, SD2, and 4D. Taken together, these common characteristics indicate that the conformational dynamics of the proteasome is evolutionarily conserved to meet its critical functional needs. Despite these shared traits in conformational dynamics of the proteasome holoenzyme, the exact structures of the RP are all somehow different, more or less, among all the states enlisted in Table 1.2, illustrating a structural plasticity of the proteasome to an exceptionally large extent, which is yet to be comparably found in other protein complexes.
Key States of Proteasome in the Act of Substrate Degradation
States EA1 and EA2 overall resemble the SA conformation of the substrate-free human proteasome holoenzyme (Dong et al. 2019). Several remarkable features distinguish states EA1 and EA2 from SA. Foremost, a ubiquitin density is observed around the T1/T2 site of RPN1 (Shi et al. 2016). Two ubiquitin densities are found at the N-terminal CC domain of RPT4-RPT5 near RPN10. The ubiquitin near RPN11 is attached to the RPT4-RPT5 CC but does not directly contact RPN11 in state EA1. This ubiquitin is bound with RPN11 and detached from the RPT4-RPT5 CC in state EA2. However, no substrate is observed inside the AAA-ATPase motor, indicating that substrate engagement is not yet completed in these states. These suggest that states EA1 and EA2 represents the steps of initial ubiquitin binding and ubiquitin transfer to RPN11, respectively, presumably before a substrate is fully engaged with the proteasome for deubiquitylation.
State EB presents a quaternary structure in the act of substrate deubiquitylation and finds no rigorous counterparts among those substrate-free proteasome conformations (Dong et al. 2019). It reveals an interesting conformation that has been absent in all other studies (Chen et al. 2016a; Huang et al. 2016; Schweitzer et al. 2016; de la Pena et al. 2018; Zhu et al. 2018). Although the overall RP-CP relationship is closer to states SA and EA, the lid swings outward away from the OB ring relative to state EA, toward an opposite direction against that seen in the EB-to-EC transition. This results in a wider access to the AAA-ATPase ring. Notably, the isopeptide bond linking substrate lysine to the C-terminal glycine of RPN11-bound ubiquitin has been observed in the vicinity to the zinc ion bound at the catalytically active site of the DUB RPN11. This DUB-substrate interaction is achieved through extensive quaternary rearrangements also involving RPT5 and RPN8. The lysine residue linking to the RPN11-bound ubiquitin via the isopeptide bond is located above the OB ring and is about 15 Å away from the central pore of the ATPase ring, where the substrate density is observed all the way from the ubiquitin-conjugated lysine down to the end of the central pore of the ATPase ring.
States EC1 and EC2 present two successive snapshots that capture the initiation steps of substrate translocation (Dong et al. 2019). While the RPN11-bound ubiquitin remains in state EC1, its isopeptide bond linking the substrate appears to have been broken. In state EC2, this ubiquitin is released from RPN11. As the overall lid-base relationship resembles the substrate-free state SC, the lid is rotated ~40° clockwise relative to states EA and EB. Although the ATPase conformation is nearly invariant in states EC1 and EC2, the ATPase ring exhibits a rigid-body translation of ~5 Å above the CP, with a small rotation in the lid. The substrate inside the AAA-ATPase ring is presumably advanced toward the CP relative to state EB. The two states exhibit similar features in the RP-CP interface with a closed CP gate.
States ED1 and ED2 present two sequential conformations in which substrate-pore loop interactions are consistent with processive substrate translocation (Dong et al. 2019). The substrate is presumably moved toward the CP relative to states EC1 and EC2. No ubiquitin densities are found on RPN11. The overall lid-base relationship, the RP-CP interface and the open state of CP gate are compatible with the substrate-free states SD1, SD2 and SD3 of the human proteasome (Chen et al. 2016a; Zhu et al. 2018), and the substrate-engaged states 5T, 4D (de la Pena et al. 2018), the tetraubiquitin-bound states C3-a, C3-b (Ding et al. 2019), and substrate-free states s4, s5 and s6 of the yeast proteasome (Wehmer et al. 2017; Eisele et al. 2018). It is noteworthy that the pore-loop staircase architecture of states 5D, 5T, C3-b and s6 closely resembles that of state ED1, whereas the pore-loop topologies of states SD2, 4D, C3-a and s4 match state ED2.
Allosteric Regulation of Proteasome Dynamics
Ligand binding and substrate interactions substantially modify the energy landscape of the proteasome and thus regulate both the distribution of coexisting conformers and the key features of these conformers (Fig. 1.9). In the presence of ATP/Mg2+, the substrate-free human proteasome exhibits four conformational states SA, SB, SC and SD in the particle populations at 76.1%, 10.2%, 5.8% and 7.9%, respectively. In the presence of ATPγS (that completely replaces ATP), the expanded states SA, SB, SC, SD1, SD2 and SD3 represent 51.8%, 3.5%, 5.3%, 14.9%, 17.0% and 7.5% of the total particles, respectively (Zhu et al. 2018). The particle population of the human proteasome in an open-gate state is about five times higher in the presence of ATPγS than ATP. Indeed, the peptide-hydrolyzing activity of the proteasome has been observed to be substantially stimulated by ATPγS (Li and Demartino 2009). In contrast to the shift of conformational equilibria toward the open-gate states, binding of the small-molecule inhibitor Oprozomib to the β5 subunit in the CP stabilizes the resting state (SA) by substantially narrowing the ground funnel on the energy landscape and inhibiting the human proteasome from sampling those states with an open CP gate (Haselbach et al. 2017).
Conceptual schematic illustrating the regulation of the energy landscape of the 26S proteasome by substrates and nucleotides. The replacement of ATP with slowly hydrolyzed ATPγS enhanced the conformational states corresponding to an open CP gate. The engagement of a substrate with the proteasome further stabilizes these states and induces more sub-states corresponding to a closed CP gate
Structural and biochemical studies on the substrate-free yeast proteasome also agree with this ligand-dependent regulatory picture in general. Binding of ATPγS or other nucleotide analogs, such as AMP-PNP, ADP-AlFx and ADP-BeFx, substantially induces ATPase conformations resembling those substrate-engaged states and shifts the conformational landscape of the proteasome from the resting state (s1) toward the open-gate states (s4, s5 and s6) (Wehmer et al. 2017; Eisele et al. 2018; Ding et al. 2017). Similar allosteric effects have also been observed for the Lys48-linked tetraubiquitin-bound yeast proteasome (Ding et al. 2019). In this case, the conformational equilibria of the proteasome are driven toward two open-gate states (C3-a and C3-b), which resemble the substrate-engaged human states ED2 and ED1, respectively (Dong et al. 2019). Notably, the resting state no longer dominates the energy landscape in some conditions. For example, 2 mM ATPγS significantly expands the particle population of state s3, s4 and s5, whereas 4 mM ATPγS even further promotes the distribution of state s3 and s4, allowing the two states to dominate the energy landscape of the yeast proteasome (Eisele et al. 2018).
Because the resolution of most yeast proteasome reconstructions was insufficient to define the nucleotide states in the proteasomal ATPases, a genetic approach has been developed to evaluate how individual ATP-binding events influence the conformational landscape of the proteasome (Eisele et al. 2018). The conserved Walker B glutamate was mutated to glutamine in each ATPase subunit in yeast, which was codenamed rpt-EQ. While mutations in three ATPases caused cytotoxic effects, mutant proteasomes from rpt2-EQ, rpt3-EQ, and rpt6-EQ enabled cryo-EM analysis that showed a consistent stimulation of states s4, s5 and even discovery of a new conformational state s6 of the yeast proteasome. While states s4 and s5 were observed in rpt2-EQ and rpt6-EQ proteasomes, state s6 was exclusively observed in rpt3-EQ proteasome, in which the ATPase conformation resembles state ED1.
In the presence of both ATP/Mg2+ and ATPγS/Mg2+, the substrate-engaged human proteasome shows seven conformers EA1, EA2, EB, EC1, EC2, ED1 and ED2 in the particle populations at 8.4%, 6.4%, 19.4%, 9.0%, 5.7%, 23.1% and 27.9%, respectively (Dong et al. 2019). The open-gate states ED1 and ED2 dominate the energy landscape, in stark contrast to the conformational distributions of substrate-free proteasome. Because all these seven states have been reconstructed at a resolution no worse than 3.6 Å, these human proteasome structures afforded unambiguous determination of the nucleotide states of the ATPase, allowing the elucidation of how the nucleotide hydrolysis and exchange trigger state transitions of the proteasome (Dong et al. 2019). Similar allosteric effects were also noted for the substrate-bound RPN11-inactivated yeast proteasome (de la Pena et al. 2018). These structural data explain why substrate engagement considerably stimulates the ATPase activity and serve as a foundation to further understand how ubiquitin-like (UBL) proteins stimulate proteasome activation (Kim and Goldberg 2018; Collins and Goldberg 2020).
In vivo experimental studies using cryo-electron tomography (cryo-ET) and subtomogram classification suggest that the 26S proteasome mostly stays at the resting state in intact hippocampal neurons in the absence of proteotoxic stress, and exhibits several alternative conformations (Asano et al. 2015; Guo et al. 2018). Although the subtomogram averaging from cryo-ET reconstructions has a low resolution compared to single-particle reconstruction, the large RP rotation between the resting state (SA) and the substrate-degrading states (ED) is discernible at a nanometer resolution. Subtomogram classification has been conducted on the proteasome within intact neurons showing poly-Gly-Ala (poly-GA) aggregates, which results from aberrant translation of an expanded GGGCCC repeat in the C9orf72 gene. Mutation in C9orf72 is the most common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Edbauer and Haass 2016; Freibaum and Taylor 2017; Gendron et al. 2013; Lin et al. 2017). The poly-GA aggregates in neurons densely pack into twisted ribbons and recruit a large number of 26S proteasomes that exist in either the substrate-degrading states (ED) or the resting state (SA) (Guo et al. 2018). The cryo-ET studies provide strong evidence supporting that the in vivo proteasome conformational dynamics regulated by the endogenous substrates are reflected in those analyzed in vitro at the atomic level.
Activation Dynamics of the CP
By studying the Thermoplasma acidophilum CP, which provides a lower structural complexity, the NMR quantification of structural dynamics of activator binding and specifically labeled residues (Sprangers and Kay 2007) elucidated the mechanism of CP gating by the unstructured N-termini of α-subunits in the archaeal proteasomes. It was observed that on average two of the chains move through the CP gate to the proteolytic chamber, and block the passage for translocating protein substrates (Religa et al. 2010). The NMR studies of the CP were conducted by methyl transverse relaxation optimized spectroscopy, which takes advantage of deuterated proteins and selectively labelling methyl groups of isoleucine, leucine, methionine and valine (Kay 2011).
How the CP gate is regulated by its activators has been informed by numerous structural studies. Crystal structure of the human PA28α/REGα homolog reveals a toroidal heptamer architecture. The CP-interacting C-terminal residues and internal activation loop exhibit sevenfold symmetry (Knowlton et al. 1997). The 20-Å pore inside this heptamer was initially suggested to be a substrate translocation channel. However, it was later found that this pore is occupied in the PA26 homolog of Trypanosoma brucei distantly related to PA28 (Forster et al. 2005). Crystal structures of PA26 in complex with the CPs in Saccharomyces cerevisiae (Whitby et al. 2000; Forster et al. 2003) and Thermoplasma acidophilum (Forster et al. 2005) indicate that the C-termini of PA26 are docked into the α-pockets, whereas the activation loops trigger the opening of the CP gate by repositioning the Pro17 turn of the CP in a sevenfold symmetric fashion.
Crystal structures of a Blm10-CP complex exhibit an architecture distinct from that of PA28. The single-chain ~250-kDa Blm10 activator binds the α-ring and resembles the shape of a turban (Iwanczyk et al. 2006; Sadre-Bazzaz et al. 2010; Schmidt et al. 2005). Interestingly, Blm10 docking onto the CP induces a disordered conformation of the CP gate. There appears to be only limited access to the turban-like architecture of Blm10 above the CP gate, consistent with a lower level of peptidase activities in the presence of Blm10 compared to PA26 (Iwanczyk et al. 2006). One C-terminal tail of PA200 docks the α5-α6 pocket, with the three C-terminal residues closely resembling the C-termini structure of PA26, forming comparable mainchain hydrogen bonds and electrostatic interactions (salt bridge) with the surface-exposed lysine in the α6 subunit. This is insufficient to open the CP gate, because other α-pockets are not occupied and conserved Blm10 residues sterically impedes the gate opening. The structures of the PA28 and Blm10/PA200 in complex with the CP point to the same binding mechanism of the CP activators in general. In both cases, a salt bridge is formed between the C-terminal carboxylate of the activator and the surface-exposed lysine of the CP (Forster et al. 2005; Dange et al. 2011; Sadre-Bazzaz et al. 2010) and the Pro17 turns of the CP is repositioned, resembling their configuration in the PA26-CP complexes. These features underscore the functional importance of the penultimate tyrosine (or phenylalanine) in the C-termini of the CP activators (Smith et al. 2007). This mechanism of CP gating has been further supported by crystal structures of the PA26 mutants binding the archaeal CP (Stadtmueller et al. 2010; Yu et al. 2010), a cryo-EM structure of the CP in complex with the PAN C-terminal peptides (Rabl et al. 2008), and high-resolution cryo-EM structures of the human 26S proteasome (Chen et al. 2016a; Zhu et al. 2018; Dong et al. 2019).
The structures of the PA28 and PA200 complexes do not intuitively clarify their elusive biological function (Rechsteiner and Hill 2005). The expression of PA28α and PA28β can be induced by interferon-γ, implicating a potential role in T-cell antigen presentation mediated by major histocompatibility complex class I (MHC-I) (Sijts et al. 2002). However, the mechanism underlying this process remains elusive, because many species that do not seem to encode MHC-I also express a PA28 homolog (Sijts and Kloetzel 2011). PA28γ/REGγ was observed to mediate ATP-independent and ubiquitin-independent degradation of some natively unstructured transcription factors (Chen et al. 2007; Li et al. 2007a). PA200 has been proposed to play a role in maintenance of mitochondrial inheritance, in DNA repair and in spermatogenesis. The abundance of the PA200-CP complexes was increased when either activated or disrupted CP prevails (Savulescu and Glickman 2011). Both PA28 and PA200 can participate in the assembly of hybrid proteasomes, in which different types of CP regulators bind to opposite sides of the same CP (Cascio et al. 2002; Kopp et al. 2001; Tanahashi et al. 2000). Such hybrid proteasomes with the PA28 or PA200 at one side and the RP at the other side have indeed been observed on the endogenously purified eukaryotic proteasome by cryo-EM in my laboratory (unpublished data). The hybrid proteasome is expected to possess distinct gating dynamics on opposite sides of the same CP. Interestingly, the gate conformations at the opposite sides of the archaeal CP appear to be coupled, which might allosterically regulate the assembly of hybrid proteasomes (Yu et al. 2020). Structural and functional studies of the hybrid proteasomes are anticipated to clarify their roles in regulating cellular processes.
In the 26S proteasome, the conformation of the CP gate is controlled by the RP-CP interface in an ATP-dependent fashion. The intermediate states of the RP-CP interface are characterized in the substrate-engaged proteasome. Perhaps the most interesting observation at this critical interface is the insertion of one additional C-terminal tail of RPT subunits into one α-pocket at a time, when the proteasome navigates from state EA, through states EB and EC to state ED. In the resting state (SA) or substrate-accepting states (EA1 and EA2), RPT3 and RPT5 have already docked their C-terminal HbYX motifs into two α-pockets (α1-α2 and α5-α6). State transition to EB is accompanied with RPT2 HbYX motif insertion into the α3-α4 pocket, followed by RPT6 C-terminus insertion into the α2-α3 pocket in State EC and RPT1 HbYX motif insertion into the α4-α5 pocket in State ED (Fig. 1.10) (Dong et al. 2019). Particularly, the RP-CP interaction modes of substrate-engaged human states EC and ED were consistently found in those of substrate-free yeast states s3 and s4-s6, respectively (Eisele et al. 2018).
Gating mechanism of the CP in the human proteasome (Chen et al. 2016a; Zhu et al. 2018; Dong et al. 2019). a Comparison of the cryo-EM densities of the RPT C-termini binding the α-pockets of the CP in different conformational states of the human proteasome. The cryo-EM reconstructions of the RP-CP interfaces are shown in grey surface representation. The colored dashed circles highlight the densities of the RPT C-terminal tails. b A schematic diagram summarizes the RPT C-terminal interaction with the α-pockets and the state of the CP gate in all states shown in (a). The ATPase ring is represented as a hexagon, the CP as a heptagon and the RPT C-terminal interaction as a colored sphere
The principal gate-blocking N-terminal loops are from the α2 and α4 subunits (Groll et al. 2000). The α3 N-terminal loop behaves as a lynchpin of the CP gate overlaid with the N-terminal loops of α2 and α4 (Tian et al. 2011). Destabilization of the α3 lynchpin is necessarily but insufficiently controlled by RPT2. By comparison, the yeast RP-CP interaction in the resting state (s1) exhibits the same mode observed in state EB of the human proteasome, in which three HbYX motifs from RPT2, RPT3 and RPT5 are inserted in the α-pockets (Wehmer et al. 2017; Dong et al. 2019). In contrast to the sequential insertion of RPT C-terminal tails in the human proteasome during the process of CP activation, the yeast CP apparently requires fewer intermediate steps in full activation (Eisele et al. 2018). It is also notable that once four RPT C-terminal tails docked into the α-pockets, the CP gate starts to show partial opening, as captured in the substrate-bound state 5D (de la Pena et al. 2018), states EC1 and EC2 (Dong et al. 2019) and substrate-free state s3 (Wehmer et al. 2017).
The insertion of five RPT C-terminal tails also demands the repositioning of the ATPase ring above the CP, as well as necessary RPT conformational rearrangements to satisfy the geometrical requirement. In states ED or SD, the N-terminal loops of α2, α3 and α4 are rotated ~90° to approximately align along the heptameric axis to open the CP gate. However, the secondary structural elements surrounding the gate-blocking loops in the α-ring remain largely unchanged with only very subtle movements relative those in other states; indeed, the overall outer diameter of the opened α-ring is dilated by only ~3 Å (Zhu et al. 2018; Dong et al. 2019). A similar allosteric effect has also been observed in the open gate of yeast CP in complex with the PA26/11S regulator from Trypanosoma brucei (Whitby et al. 2000; Stadtmueller et al. 2010). Importantly, in both the RP-activated human CP and the PA26-activated yeast CP, the same α-pockets (α6-α7 and α7-α1) are the only two that are empty, indicating a highly conserved mechanism of CP gating regulation by its activators.
The CP is not only externally regulated by the RP, but is also internally affected by the interactions at the proteolytic sites in the β-type subunits (Haselbach et al. 2017; Osmulski et al. 2009). Studies of the yeast CP in the absence of RP using atomic force microscopy have suggested that the CP gating is either stimulated or suppressed by the proteolytic active-site engagement with a substrate or small-molecule inhibitors such as Bortezomib and Epoxomicin (Osmulski et al. 2009). The CP gate opening appears to be stimulated by a tetrahedral transition state at the proteolytic active sites. Notably, such a short-range allosteric effect on the CP gate can be further transmitted to the conformational state of RP through the RP-CP interface in a long-range fashion (Haselbach et al. 2017).
Quaternary Allosteric Regulation
The activity of proteasomal ATPase motor is regulated by the lid subunits, RPN1 and RPN2 in an asymmetric fashion (Greene et al. 2019). While in states ED1 and ED2, RPN1 and RPN2 form a dimer by a long-range association through a long turning loop from RPN2, such a quaternary architecture was less clearly observed in other states (EA-C) (Dong et al. 2019). During state transition from EA to EB, RPN1 undergoes a prominent rotation. In states EC1 and EC2, the RPN1 density is blurred, indicating strong motions potentially disrupting the RPN1-RPN2 associations observed in states ED1 and ED2. Thus, the variation of RPN1 motion appears to be highly coordinated with the ATP hydrolytic cycle coupled with conformational remodeling of the ATPase motor during substrate translocation.
The CC domains of RPT4-RPT5 also seem to coordinate substrate translocation by its interaction with the lid subunit. In states EC1, EC2, ED1 and ED2, the RPT4-RPT5 CC domain interacts with either RPN9 or RPN10 in different contacts. By contrast, it resides several nanometer away from any RPN subunit and recruits peptide-proximal ubiquitin in state EA and transfers it to RPN11 for deubiquitylation in state EB. Thus, the lid-base interaction regulates substrate interactions with the proteasome through a combination of short-range and long-range allosteric regulations, which are supported by a recent biochemical study (Snoberger et al. 2018). The conformational regulation between the lid and base or CP is similarly observed in the absence of substrates (Zhu et al. 2018).
The lateral RP-CP interface, particularly between the RPN6 PCI domain and the α2 subunit, also demonstrates prominent fluctuations that interconvert between open and closed configurations. This interfacial instability is more prominently observed in the states corresponding to an open CP gate or in the presence of substrates, indicating an ancillary role of this interface in the regulation of the conformational changes of the axial substrate-translocation pathway (Zhu et al. 2018; Dong et al. 2019). Indeed, the interaction of RPT6 with the α2 subunit is regulated by phosphorylation on RPT6, which stimulates proteasome activity (Satoh et al. 2001; Asai et al. 2009)
Summary of Proteasome Dynamics
In this section, complex dynamics of the proteasome in the act of substrate processing at the atomic level are compared in detail with structural characterization of proteasome dynamics at the near-atomic or pseudo-atomic level under various in vitro biochemical conditions. This retrospective comparison on 29 conformational states of the proteasome not only reveals the common traits of proteasome dynamics, but also makes clear several outstanding revelations that were relatively weak or less obvious in single studies, as summarized in the following bullet points.
-
The resting state is least active in its CP in the presence of ATP. Replacing ATP with other nucleotides or nucleotide analogs, the presence of ubiquitin and UBL proteins, or the engagement of substrates generally re-equilibrates the proteasome conformations toward the open-CP states and stimulates proteasome activation.
-
Both the RPN11-OB interface and the CP gate controlling the substrate entrance are approximately bistable. Their state transitions are desynchronized to coordinate the timing between deubiquitylation and translocation events.
-
The lid rocking is generally observed to regulate both the ATPase and CP conformations. It becomes increasingly apparent that the lid interactions with the base and CP favor two sets of ATPase conformations characterized in two sets of comparable states [ED1, 5D/5T, C3-b, s6] and [ED2, SD2, 4D, C3-a, s4]. (See below: section Operating Principles of Proteasomal AAA-ATPase Motor, for more details).
-
Substrate engagement stabilizes the states with the open CP gate, and via its interactions with the proteasome at multiple sites along the substrate-translocation pathway, coordinates the actions of ubiquitin recognition, deubiquitylation, initiation of unfolding and processive translocation through the ATPase ring and the CP gate. Such a multi-enzymatic coordination is allosterically assisted by the lid subcomplex interacting with the base and CP at multiple peripheral sites.
-
The dynamic CP gate is regulated both externally and internally via allosteric effects. This enables the CP gating to be highly responsive to inter-subcomplex coordination for processive substrate degradation without generating proteolytic errors.
-
The allosteric regulation between RP and CP is bidirectional via both short-range and long-range pathways. While the interactions of ubiquitin receptors with ubiquitin or UBLs can allosterically impact the CP state, the interaction of the CP proteolytic active sites with substrates or inhibitors can also influence the RP state. Once the RP and CP are assembled into a holoenzyme, the conformational dynamics of all subunits are allosterically, coherently, and globally coupled. No single subunit acts alone without triggering coherent allosteric effects within the holoenzyme.
Life Cycle of the Proteasome
Proteasome Biogenesis
Constitutive Proteasome Expression
The cellular abundance of the proteasome is dynamically regulated by a genetic circuit of negative feedback at the transcriptional level (Motosugi and Murata 2019) (Fig. 1.11). Specific transcription factors regulating expression of the proteasome subunits have been identified (Budenholzer et al. 2017; Rousseau and Bertolotti 2018). In Saccharomyces cerevisiae, a transcription factor Rpn4 control coordinated gene expression of proteasome subunits (Wang et al. 2008). Rpn4 is a zinc-finger protein and binds a conserved sequence motif (5′-GGTGGCAAA-3′) known as proteasome-associated control element (PACE) in the promoter region of genes expressing proteasomal subunits and assembly chaperones (Mannhaupt et al. 1999; Shirozu et al. 2015). Rpn4 is rapidly degraded by the proteasome and is thus short-lived (t1/2 ≈ 2 min). Through such a negative feedback circuit, Rpn4 promotes proteasome expression whenever the proteasome function is compromised (Xie and Varshavsky 2001). Loss of RPN4 gene or PACE sequences compromises proteasome activity and cell viability under various stress conditions such as oxidative DNA damage (Wang et al. 2008). Rpn4 abundance is also regulated by many stress-inducible transcription factors, such as Hsf1, Pdr1, Pdr3, and Yap1 (Owsianik et al. 2002; Ma and Liu 2010). Increasing proteasome expression might be a common mechanism for cells to adapt to diverse challenging conditions (Hahn et al. 2006).
Life cycle of the 26S proteasome. Biosynthesis of proteasome subunits from individual amino acids is regulated by transcription factors such as NRF1 in mammals, NAC53/NAC78 in plants and Rpn4 in yeast. With assistance of CP and RP assembly chaperones, the expressed subunits assemble into the mature form of the 26S proteasome in a coordinated manner. Proteasomes are localized in either the nucleus or cytosol, where their activities can be regulated by numerous proteasome-associated proteins and by post-translational modifications. Extrinsic and intrinsic ubiquitin receptors recognize and deliver polyubiquitylated substrates to the proteasome for degradation. Damaged proteasomes are degraded in the lysosome or vacuole via autophagy, which is mediated by signals from the nutrient-responsive ATG1 kinase, subunit ubiquitylation, and several autophagy receptors, including p62/SQSTM1 in mammals, RPN10 in plants, and Cue5 in yeast
Although mammalian cells lack of any Rpn4 orthologs, several transcription factors regulating the mammalian proteasome expression have been identified. One of such transcription factors is nuclear factor erythroid-derived 2-related factor 1 (NFE2L1) also named NRF1 (Radhakrishnan et al. 2010). NRF1 promotes expression of all genes encoding proteasome subunits and their assembly chaperones in the presence of proteasomal dysfunction, leading to de novo proteasome synthesis. In Arabidopsis thaliana, the transcription factors NAM/ATAF1/CUC2 78 (NAC78) and NAC53 were proposed to play a similar role as Rpn4 and NRF1 in upregulating proteasome synthesis that help plants survive proteotoxic stress (Gladman et al. 2016).
In contrast to NRF1, several transcription factors specifically regulate the induction of a portion of proteasome subunits in mammalian cells. Nuclear transcription factor Y (NF-Y) is a complex consisting of three proteins, NF-YA, NF-YB, and NF-YC. NF-Y recognizes the CCAAT motif in the promoter region of certain proteasomal genes and upregulates cellular proteasome activity. The CCAAT motif is found in the genes of six CP subunits (α2, α5, α7, β3, β4, and β6), five RP subunits (RPT1, RPT5, RPT6, RPN10, and RPN11) and one RP assembly chaperone (p28) (Xu et al. 2012). Other transcription factors, such as the forkhead box protein O4 (FOXO4) and the signal transducer and activator of transcription 3 (STAT3), exhibit seemingly greater specificity in the regulation of proteasome subunit expression. FOXO4 specifically regulates RPN6 expression and maintains the proteasome activity at a higher level in human embryonic stem cells (Vilchez et al. 2012; Webb and Brunet 2014). In the JAK/STAT pathway, JAK phosphorylation upon cytokine signaling activates STAT3, which upregulates PSMB5 (β5 subunit) expression through epidermal growth factor (EGF) induction (Vangala et al. 2014).
Proteasome abundance is also regulated at the post-transcriptional or translational level (Rousseau and Bertolotti 2016; Kors et al. 2019; Motosugi and Murata 2019). In yeast, the mitogen-activated protein kinase 1 (Mpk1) regulates expression of the proteasome subunits and RP assembly chaperones (RACs) under stress conditions or in response to rapamycin and tunicamycin. However, mRNA levels are comparable between wild-type and Mpk1-deficient cells, suggesting that Mpk1-mediated regulation of the proteasome occurs at the translational level. Moreover, the translation of RPT5 mRNA can be suppressed by haploinsufficiency of ribosomal genes, which primarily causes Diamond-Blackfan anemia (Khajuria et al. 2018). In hematopoietic stem and progenitor cells, translation of proteasome mRNA is selectively inhibited by the storage of ribosomes, thus impeding erythroid lineage commitment.
The growth-controlling kinase mTORC1 (Mammalian or mechanistic target of rapamycin complex 1) is activated in response to growth factors or increased nutrient levels. It activates protein translation and lipid synthesis to promote cell growth and proliferation (Dibble and Manning 2013). The mTORC1 pathway has been found to control proteasome abundance at multiple levels (Zhang et al. 2014; Zhang and Manning 2015; Rousseau and Bertolotti 2016; Zhao et al. 2015). The inhibition of mTORC1 can induce ERK5, the human orthologs of Mpk1, which upregulates the RACs and proteasome subunits (Rousseau and Bertolotti 2016). Stimulated by growth factor signaling, mTORC1 can activate sterol regulatory element binding protein (SREBP-1) that regulates expression of lipogenic genes (Ricoult and Manning 2013). The mTORC1-activated SREBP-1 is related to NRF1 expression that regulates proteasome expression (Zhang et al. 2014). On the other hand, mTORC1 also phosphorylates and inactivates ATG proteins required for autophagy induction, such as ATG1 and ATG13 (Hosokawa et al. 2009; Dibble and Manning 2013). Since the proteasome is destructed by autophagy (Marshall and Vierstra 2015; Marshall et al. 2015; Marshall et al. 2016; Cohen-Kaplan et al. 2016; Waite et al. 2016; Nemec et al. 2017), mTORC1 regulates both the proteasome biosynthesis and breakdown in response to nutrient conditions.
Immunoproteasome Expression
Proteasomal subtypes are differentially expressed in response to certain cellular conditions to meet specific functional needs (Motosugi and Murata 2019; Ferrington and Gregerson 2012). Immune cells constitutively express the immunoproteasomes at high levels. The immunoproteasome primarily processes antigens for their presentation in the MHC-I pathway by degrading the antigens into short polypeptides. The transporter associated with antigen processing (TAP) complex, a TAP1-TAP2 heterodimer, then transports the antigen polypeptides to the ER, where the MHC-I molecules recognize the polypeptides for antigen presentation on the cell surface (Leone et al. 2013). The three catalytic subunits β1, β2, and β5 are replaced by subtype subunit β1i, β2i, and β5i, respectively, in the immunoproteasome (Kloetzel 2001). While PSMB10 (β2i) is located outside the MHC locus, PSMB9 (β1i) and PSMB8 (β5i) resides in the MHC-II region next to the genes encoding TAP1-TAP2.
The expressions of β1i, β2i, and β5i are induced by interferon-γ in response to oxidative stress or inflammatory stimuli (Aki et al. 1994; Hallermalm et al. 2001; Hussong et al. 2010; Hisamatsu et al. 1996). Interferon-γ induces protein expression involved in antigen presentation, including the proteasome activator PA28α/β, and TAP1/TAP2 (Ma et al. 1997; Realini et al. 1994; Ahn et al. 1995). β1i expression requires the GC boxes and the transcription factor SP1 (Wright et al. 1995). The genes encoding β1i and TAP1 share a bidirectional promoter that lacks any TATA box and includes several GC boxes, interferon consensus sequence 2 and γ-interferon-activated sequence sites, which are recognized by the interferon regulatory factor 1 (IRF1) and STAT1 (Wright et al. 1995; Chatterjee-Kishore et al. 2000). β1i expression is induced by IRF1 and STAT1 under interferon-γ stimulation. The nuclear factor-κB (NF-κB), a family of transcription factors, is required for the tumor necrosis factor-α (TNF-α) dependent induction (Wright et al. 1995). The β2i promoter contains NF-κB, IRF1 and SP1 consensus sequences that lack the CAAT or TATA boxes (Cruz et al. 1997; Hayashi et al. 1997). The β5i promoter contains an NF-κB consensus sequence, a GC-rich region and a TATA box (Zanelli et al. 1993). In neurons, expression of β1i and β5i is induced by the transcription factor Zif268 (James et al. 2006).
Thymoproteasome Expression
Thymic cortex specifically expresses the thymoproteasome, another proteasome subtype that plays an important role in the selection of CD8+ T cells (Murata et al. 2007; Murata et al. 2018). In the thymic cortex and medulla, immature thymocytes are subject to positive and negative selection, respectively. In contrast to the immunoproteasomes expressed in medullary thymic epithelial cells (mTECs), the thymoproteasomes is assembled from the subtype subunit β5t in replacement of β5 and is specifically expressed in cortical thymic epithelial cells (cTECs). The PSMB11 gene encoding β5t is adjacent to PSMB5 encoding β5. The gene product is encoded by a single exon in both the human and mouse genomes. The forkhead transcription factor FOXN1 promotes both TEC lineage specification and β5t expression (Romano et al. 2013; Uddin et al. 2017). The β5t promoter contains the conserved FOXN1-binding sequence 5′-ACGC-3′, in which a single mutation impairs CD8+ T cell production and β5t expression. Paradoxically, because FOXN1 is present in mTECs that do not express β5t at any detectable level, FOXN1 alone does not sufficiently regulate β5t expression, suggesting that other unknown cellular factors might contribute to the induction of β5t expression.
Proteasome Assembly
CP Assembly Chaperones
The assembly of the eukaryotic CP is more complex than their prokaryotic counterparts and requires a set of extrinsic and intrinsic chaperones (Table 1.1) (Murata et al. 2009; Tanaka 2009; Rousseau and Bertolotti 2018; Budenholzer et al. 2017). The complex process of CP assembly can be understood as three consecutive steps in eukaryotes (Fig. 1.12a) (Murata et al. 2009; Tomko and Hochstrasser 2013; Rousseau and Bertolotti 2018): (1) α-ring formation, (2) β-ring formation and (3) half-CP dimerization and proteasome maturation. These intermediate assembling steps are assisted by five proteasome chaperones named proteasome assembling chaperone-1 (PAC1)-PAC4 and proteasome maturation protein (POMP) in human (Witt et al. 2000; Murata et al. 2009). The yeast orthologs of PAC1-PAC4 and POMP are known as proteasome biogenesis-associated 1 (Pba1)-Pba4 and underpinning maturation of proteasome 1 (Ump1), respectively.
Models of the RP and CP assembly in eukaryotic cells. a Assembly pathway of the constitutive CP. The α-ring is assembled with the help of two heterodimers of chaperones: PAC1-PAC2 and PAC3-PAC4. β2 and POMP then associate with the α-ring. The incorporation of β3 and β4 coincides with the dissociation of PAC3-PAC4, followed by sequential incorporation of β5, β6, β1 and β7. Dimerization of two half-CP forms the preholoproteasome. In the last step, the propeptides of the β-ring is auto-cleaved to activate the CP; POMP and PAC1-PAC2 are then degraded to form the mature CP. b Hypothetical assembly pathway of the RP. The base assembly is assisted by four RACs: S5b, PAAF1, p27 and p28. The lid can be assembled via a hierarchical pathway without any chaperones. It starts with the formation of two subcomplexes Module 1 and LP3. They associate to form LP2. The final step occurs with RPN12 joining LP2 to form the mature lid. The lid then binds the free base. Note that in yeast the base may first assemble with the CP before its association with the lid
Four CP assembly chaperones guide the assembly procedure of the α-ring. They form two heterodimers, PAC1-PAC2 (Hirano et al. 2005) and PAC3-PAC4 in human (Pba1-Pba2 and Pba3-Pba4 in yeast) (Hirano et al. 2006; Le Tallec et al. 2007). The PAC1-PAC2 heterodimer promotes the assembly of the α-ring intermediate and prevents premature binding of the RP to the α-ring, as well as aberrant dimerization of α-rings (Hirano et al. 2005; Wani et al. 2015; Stadtmueller et al. 2012; Kusmierczyk and Hochstrasser 2008). Knockdown of PAC1 or PAC2 expression decreases the amount of α-rings and results in accumulation of off-pathway products of aberrant α-ring dimers. PAC1-PAC2 binds to proteasome precursors until the complete formation of the CP. PAC1-PAC2 is degraded by the newly assembled CP and has a half-time of about half an hour (t1/2 ≈ 30 min), which is consistent with the maturation time of the CP.
In yeast, the Pba1-Pba2 heterodimer ensures the appropriate incorporation of the α5 and α6 subunits, whereas the Pba3-Pba4 heterodimer interacts with the α5 subunit of the CP intermediate to ensure the appropriate assembly. Crystal structure of the Pba3-Pba4-α5 complex exhibits pronounced similarity to the structure of the PAC3 homodimer (Yashiroda et al. 2008). Notably, the structures of Pba3 and Pba4 are mostly homologous to the CP subunits. Structural analysis of the Pba3-Pba4-α5 complex suggests that Pba3-Pba4 binds the facet of the α-ring opposite that contacted by Pba1-Pba2, thus sterically clashing with the incoming β4 subunit. This explains why Pba3-Pba4 has to dissociate as β-subunits are incorporated. Deletion of the Pba3 or Pba4 gene causes the accumulation of proteasome intermediates and produces diverse aberrant CP, including those lack of α4 or harboring a second copy of α4 instead of α3 (Velichutina et al. 2004; Kusmierczyk and Hochstrasser 2008; Padmanabhan et al. 2016). Thus, Pba3-Pba4 promotes and guides the proper assembly of the α-subunits, especially α3 and α4.
The α-ring serves as a template for the assembly of the β-ring, which begins with the sequential incorporation of β2, β3 and β4 subunits. POMP/Ump1 is incorporated along with the first β-subunits, whereas the PAC3-PAC4/Pba3-Pba4 dimer is released from the α-ring upon β3 integration (Hirano et al. 2006; Hirano et al. 2008; Li et al. 2007b). The resulting intermediate, known as the 13S complex, then progressively recruits β5, β6, β1 and β7 to form a half CP assembly, also named the 15S complex. Half-CP dimerization is immediately initiated after β7 incorporation (Li et al. 2016b).
The final stage of joining two half CP assemblies together is regulated by POMP/Ump1. POMP prevents premature dimerization of half CP by hindering their dimerization until all β-subunits are incorporated properly (Ramos et al. 1998). After half CP dimerization, the N-terminal propeptides from the β1, β2, β5, β6 and β7 subunits are cleaved in the nascent CP for maturation. Human POMP and PAC1-PAC2 are then degraded by the newly assembled proteasome (Hirano et al. 2006), whereas yeast Pba1-Pba2 is recycled for assembly of newly synthesized CP subunits in yeast (Ramos et al. 1998; Kock et al. 2015). Although there is lack of structural information for POMP/Ump1 interactions, crystal structures of the Rhodococcus proteasome mutants with its intact propeptides have offered insights into the final step of CP maturation (Kwon et al. 2004; Witt et al. 2006). These structures show that the propeptide contacts two adjacent α-subunits, thereby promoting CP assembly and maturation.
RP Assembly Chaperones (RACs)
The lid and base subcomplexes of the RP can assemble separately without and with the RACs, respectively (Fig. 1.12) (Isono et al. 2007). Recombinant lid subunits can self-assemble into a correct subcomplex in the absence of the CP or the base subcomplex in vitro (Lander et al. 2012; Tomko and Hochstrasser 2014). The lid assembly in yeast is preceded by the formation of two intermediate complexes, one named LP3 containing Rpn3, Rpn7 and Sem1, and the other named Module 1 composed of Rpn5, Rpn6, Rpn8, Rpn9 and Rpn11 (Tomko and Hochstrasser 2014; Fukunaga et al. 2010). The intermediate subassemblies LP3 and Module 1 associate to form the subassembly LP2 through the formation of the C-terminal helical bundle, which is finalized by the incorporation of the lid subunit RPN12 (Estrin et al. 2013; Tomko et al. 2015). It remains to be clarified if there are unknown RACs assisting the lid assembly in vivo. The pathway of lid assembly in mammalian cells is not well understood.
Assembly of the base subcomplex is regulated by four RACs in mammalian cells (Table 1.1): S5b (Roelofs et al. 2009; Kaneko et al. 2009), p28 (gankyrin) (Krzywda et al. 2004; Nakamura et al. 2007a), PAAF1 (proteasomal ATPase-associated factor 1) and p27 (Park et al. 2010). Their yeast orthologs are Hsm3, Nas6, Rpn14 and Nas2, respectively. The base assembly is centered around the formation of the heterohexameric ring of AAA-ATPases (RPT1-RPT6). Each RP assembly chaperone binds the C-terminal domain of a distinct RPT subunit in pairs of S5b-RPT1, p28-RPT3 and p27-RPT5, forming three intermediate complexes S5b-RPT1-RPT2-RPN1, p28-RPT3-RPT6-PAAF1 and p27-RPT5-RPT4, respectively (Kaneko et al. 2009; Park et al. 2009; Roelofs et al. 2009; Thompson et al. 2009; Tomko and Hochstrasser 2013; Saeki et al. 2009b). In yeast, an intermediate in base assembly named BP1 has been identified, which contains three Rpts, Rpn1, and Hsm3 (Park et al. 2009). Despite their functional similarity, the four base RACs adopt different structures. Crystal structures reveal that S5b/Hsm3 comprises HEAT repeats (Takagi et al. 2012), that p28/Nas6 comprises ankyrin repeats (Nakamura et al. 2007a), and that PAAF1/Rpn14 forms a WD40 propeller (Kim et al. 2010). The p27/Nas2 sequence predicts a PDZ domain. Modeling of the crystal structure of yeast Hsm3-Rpt1 and Nas6-Rpt3 complexes (Nakamura et al. 2007a, 2007b; Takagi et al. 2012) into the proteasomal ATPase ring suggests that Hsm3 and Nas6 physically occlude the formation of proper RP-CP contacts.
Mechanism of RP-CP Association
There remain two possible assembly pathways for the proteasome holoenzyme. The first pathway is that the CP templates the assembly of the base and the CP-base assembly intermediate further templates the lid assembly for the completion of the 26S assembly. This pathway might dominate in yeast cells, given evidence of observing the base-CP assembly intermediates (Park et al. 2009; Park et al. 2013). The second pathway is that the lid and base assembled into a free RP intermediate, which then assembles with the CP into the holoenzyme. This pathway might dominate in mammalian cells, because the assembly intermediates of the lid with a partial base have been observed in mammalian cells and there is lack of evidence on the base-CP subassemblies in these studies (Lu et al. 2017b; Thompson et al. 2009). Whether the two distinct assembly pathways can coexist and work in parallel in the same system will require further investigations.
Cryo-EM structures of an endogenously purified human p28-RP complex provide important insights into the mechanism of p28/Nas6 in assisting the last step of proteasome assembly and maturation prior to the RP-CP association (Lu et al. 2017b). Surprisingly, the isolated endogenous p28-RP complex samples a very wide conformational landscape. Pronounced conformational dynamics of the complex have been captured by cryo-EM in seven intermediate states designated TA1 to TA7, in which the RPT subunits, RPN1 and p28 were reconstructed to subnanometer resolutions whereas the remaining components of RP were solved at 4.6 Å resolution (Lu et al. 2017b). The AAA domains of RPT subunits form a lock-washer-like shape with an opening between RPT2 and RPT6 in all but states TA6 and TA7. The opening width is variant in each state, revealing a continuum of conformational changes during an open-to-closed transition of the AAA ring (Fig. 1.12). Remarkably, p28 strongly clashes with the α2 subunit in all conformational states except TA7 when the p28-RP is docked into the atomic model of the 26S proteasome. This analysis reveals a functional role of p28 in guiding the CP to select a specific conformational state TA7 of the RP to facilitate the last step of proteasome assembly by increasing interfacial complementarity while introducing no significant occlusion. This elegant mechanism is supported by several functional and genetic studies of yeast Nas6 in the RP assembly (Li et al. 2017; Nemec et al. 2019; Park et al. 2013).
A marked structural feature in the free p28-RP lies in the OB ring being blocked by RPN11 that appears to be even tighter than that in states ED and SD of the mature proteasome (Lu et al. 2017b). This quaternary arrangement prevents substrates from being prematurely recruited by the AAA unfoldase. Comparison of the p28-RP structure with that of the mature proteasome in the resting state reveals marked conformational changes of the lid upon RP-CP association (Chen et al. 2016a; Lu et al. 2017b). The lid is rotated ~40° around the heptameric axis of the CP. As a consequence, the RPN7 PCI domain is translated ~15 Å towards the CP with repositioning of RPT3 and RPT6, whereas the N-terminal helical repeat region of the RPN6 PCI domain undergoes a prominent rotation of 40° to allow the association of RPN6 with the α2 subunit.
Structural and functional studies have unraveled a conformation-selective, enthalpy-driven mechanism for p28 release upon RP-CP association (Fig. 1.13) (Lu et al. 2017b; Li et al. 2017; Nemec et al. 2019). The presence of p28 facilitates the selection of one p28-RP conformation (TA7) by the CP to form a first-encounter complex of p28-RP-CP, whereas other p28-RP conformations prohibit RP-CP association. The first-encounter complex p28-RP-CP is transient and undergoes pronounced remodeling driven by enthalpic gain at inter-subunit and inter-domain interfaces. The gap closure between RPT3 and RPT4, which stabilized the ATPase hexamer, and the stronger interaction of the ATPases with the α-ring all contribute to the minimization of the free energy of RP-CP association, which allosterically drives p28 eviction from RPT3 (Lu et al. 2017b). Specifically, RPT3 is translated ~15 Å in order to tighten its association with RPT4, to dock its C-terminal HbYX motif into the α-pocket, and to reposition its p28-binding site toward the α1-α2 surface for p28 ejection. Consistent with this picture, a yeast proteasome mutant rpt3-Δ1, in which the C-terminal residue from Rpt3 was deleted, was observed to be unable to release Nas6 (Park et al. 2009), because the mutated Rpt3 loses the affinity of its C-terminal tail with the α-pocket and is unable to complete the Rpt3 remodeling that ejects Nas6. Although there is no structural information regarding other RACs in complex with intermediate base assembly, it is conceivable that this intriguing conformation-selective, enthalpy-driven model might similarly underlie the action mechanism of other RACs.
The mechanism of RP-CP assembly. a Two possible pathways of RP-CP assembly. b A proposed model of the p28 chaperone-guided conformational selection for RP recognition by the CP for proteasome holoenzyme assembly (Lu et al. 2017b). The schematic illustrates the hypothetical CP-independent assembly pathway of the RP prior to the 26S formation. However, this hypothesis is subject to further tests and does not exclude alternative RP assembly pathways (Park et al. 2009; Roelofs et al. 2009)
Cellular Proteasome Regulation
Localization of Proteasome
The cellular proteasome is not only highly dynamic in structure but also in composition and location (Marshall and Vierstra 2019). It could dissociate into free RP and CP subcomplexes and shuttle between the nucleus and cytosol under different physiological conditions during growth, development or proteotoxic stress. Using the fluorescent microscopy, it has been visualized in mammals, plants and yeast that the free RP and CP diffuse throughout both the compartments of nucleus and cytosol (Reits et al. 1997; Enenkel et al. 1998; Russell et al. 1999; Brooks et al. 2000; Pack et al. 2014; Marshall et al. 2015; Gan et al. 2019). In different compartments, the measured proteasome activities exhibit a large variation (Gardner et al. 2005; Chen and Madura 2014; Dang et al. 2016). The local concentration of the 26S proteasome is 830–980 nM in the nucleus of actively dividing yeast cells, but only 140–200 nM in the cytoplasm measured by fluorescence correlation spectroscopy (Pack et al. 2014). Similar results were observed in mammalian neuronal cells (Asano et al. 2015).
Transport of the proteasome between the nucleus and the cytosol could be challenging through the nuclear pores (Beck and Hurt 2017). It was suggested that the nuclear proteasome may dissociate into free CP and RP subcomplexes to enable their exportation separately (Nemec et al. 2017). In proliferating yeast, CP and RP assembly intermediates, each of which carries certain nuclear localization signals (NLS), are imported into the nucleus (Tanaka et al. 1990; Nederlof et al. 1995). An importin-α/β heterodimer comprising Srp1/Kap60 and Kap95, which are two members of the β-karyopherin family, can recognize the NLS carried by a small portion of proteasome subunits and facilitate the transportation (Enenkel et al. 1995). It has been suggested that the CP, lid and base subcomplexes are transported to the nucleus separately by the importin-α/β and the final steps of proteasome assembly occur in the nucleus (Lehmann et al. 2002; Wendler et al. 2004; Isono et al. 2007). The CP or its assembly intermediates might exist in either import-incompetent or import-competent states, depending on solvent-accessibility of the NLS motifs in certain α-subunits (Tanaka et al. 1990). In support of this hypothesis, cryo-EM studies have shown that NLS sequences are exposed on CP assembly intermediates (Kock et al. 2015; Wani et al. 2015), but are concealed in the mature proteasomes. Import of the base subcomplex into the nucleus was suggested to be mediated by an NLS in RPN2 or RPT2 (Wendler et al. 2004; Isono et al. 2007; Savulescu et al. 2011; Weberruss et al. 2013).
The proteasomes are exported from the nucleus and reversibly sequestered into cytoplasmic proteasome storage granules (PSGs) in quiescent cells (Bingol and Schuman 2006; Laporte et al. 2008; Yedidi et al. 2016; Gu et al. 2017; Marshall and Vierstra 2018b). Supply of a fresh carbon source reverses this process by stimulating rapid import of the free RP and CP back into the nucleus. Blm10 was found to facilitate nuclear import of the mature CP dissociated from PSGs, when cells restart growth from quiescence (Weberruss et al. 2013). In mouse embryonic fibroblasts, the proteasomes of over 3 days old were also observed to be localized in the cytosolic compartment (Tomita et al. 2019). In the green alga Chlamydomonas reinhardtii, cryo-ET imaging has found that the proteasomes are enriched at the inner nuclear membrane with a local concentration of over 8 μM around the nuclear pore complexes (Enenkel et al. 1998; Takeda and Yanagida 2005; Albert et al. 2017). Curiously, after neuronal stimulation, a plasma membrane-associated CP was found to specifically degrade ribosome-associated nascent polypeptides associated with the ribosome in a ubiquitin-independent fashion (Ramachandran and Margolis 2017; Ramachandran et al. 2018).
Several studies found that the proteasome can also fulfill nuclear transportation without disassembly (Reits et al. 1997; Chen et al. 2011; Savulescu et al. 2011; Pack et al. 2014), as the size of the nuclear pore complex is large enough to allow free transportation of macromolecular complexes with a diameter of up to 39 nm (Pante and Kann 2002; Burcoglu et al. 2015). To examine this conception, a 26S proteasome is genetically modified by translationally fusing the α4 subunit to RPT1 or RPT2 to stabilize RP-CP association, which did not result in detectable structural defects in the reengineered proteasome. Such a stabilized variant of the 26S proteasome appears to be normally distributed in the nucleus (Laporte et al. 2008; Pack et al. 2014). Because protein synthesis is paused during quiescence, CP intermediates would be absent for nuclear import. This suggests a transport pathway in which the older, stabilized 26S proteasomes are directly imported to the nuclear (Pack et al. 2014).
Post-translational Modifications of Proteasome
Post-translational modifications regulate proteasome assembly, abundance, localization and activity. In total, there are about 350 post-translational modifications detected in the 26S proteasome. These can be categorized into 11 different types of modifications: phosphorylation, N-acetylation, N-methylation, N-myristoylation, poly(ADP-ribosyl)ation, O-glycosylation, oxidation, sumoylation, ubiquitylation, succinylation and proteolytic truncation (Kikuchi et al. 2010; Cui et al. 2014; Hirano et al. 2016; Zong et al. 2014). These modifications provide complex layers of functional regulation that endows the proteasome with exceptional adaptability to many conditions.
Catalyzed by proteasome-associated phosphatases or kinases, phosphorylation is a more extensively studied post-translational modification in the proteasome, and affects many proteasome subunits (Iwafune et al. 2002; Lu et al. 2008; Kikuchi et al. 2010; VerPlank and Goldberg 2017; Liu et al. 2020; VerPlank et al. 2019). The proteasome disassembles into the free CP and RP after treatment with alkaline phosphatase (Satoh et al. 2001). The protein kinase A (PKA) catalyzes phosphorylation of Ser120 in RPT6, which modulates the association of RPT6 and the α2 subunit and can be dephosphorylated by the protein phosphatase 1γ (PP1γ) (Satoh et al. 2001; Asai et al. 2009). Phosphorylation of RPN6 at Ser14 by PKA stimulates multiple proteasomal activities, including peptide hydrolysis by the CP and ATP hydrolysis by the RP, leading to an overall enhanced degradation rate (Lokireddy et al. 2015; VerPlank et al. 2019). The phosphatase UBLCP1 dephosphorylates Ser361 of RPN1 by binding its UBL domain to RPN1, which controls nuclear proteasome assembly by modifying the RP-CP interaction (Guo et al. 2011; Sun et al. 2017; Liu et al. 2020). Similarly, phosphorylation of the α7 subunit regulates the association of Ecm29 with the proteasome (Wani et al. 2016).
Ubiquitylation of the proteasome subunits themselves also provides versatile ways to regulate the proteasome function. For instances, in Saccharomyces cerevisiae and Arabidopsis, extensive ubiquitylation of the inactive proteasome targets itself for autophagic clearance (Marshall et al. 2015; Cohen-Kaplan et al. 2016). Ubiquitylation of RPT5 seems to regulate an intermediate checkpoint during the base assembly (Fu et al. 2018). Ubiquitylation of the ubiquitin receptors RPN10 and RPN13 downregulates their substrate-binding affinity or orchestrates their association with UBL-UBA receptors (Isasa et al. 2010; Lipinszki et al. 2012; Jacobson et al. 2014; Zuin et al. 2015).
Although numerous studies have started revealing their potential importance in functional regulation of the proteasome, the exact function and mechanism remain unknown for the majority of these post-translational modifications. Besides phosphorylation and ubiquitylation of the proteasome that have been subject to more studies than others as mentioned above, a few additional examples of these efforts are briefly summarized in the following. N-myristoylation of RPT2 can tether the proteasome to membrane surfaces (Shibahara et al. 2002; Gomes et al. 2006; Kimura et al. 2012; Kimura et al. 2016). Attachment of O-linked N-acetylgalactosamine to RPT2 inhibits the activity of the AAA-ATPase motor and reduces the overall proteasome activity (Zhang et al. 2003). In yeast, methylation of the Rpt1 N-terminus is linked to proteotoxic stress induced by the amino acid analog canavanine or hydrogen peroxide (Kimura et al. 2013). S-glutathionylation of the α5 subunit appears to regulate the yeast CP gating (Demasi et al. 2003; Silva et al. 2012). The NatB complex catalyzes N-acetylation of the proteasome subunits that regulates proteasome localization in assembling PSGs and cell fitness during aging (van Deventer et al. 2015; Marshall and Vierstra 2018b).
Proteasome-Associated Proteins
Proteasome functions are also regulated by numerous cellular factors that are transiently or reversibly associated with the 26S proteasome (Verma et al. 2000; Wang et al. 2007; Guerrero et al. 2006). These proteins are referred to as proteasome-associated proteins and can be categorized into two groups (Tanaka 2009; Finley 2009). The first group contains proteins related to the ubiquitylation pathway. The extrinsic DUBs USP14 and UCH37 can be recruited to the proteasome by association with RPN1 and RPN13, respectively (Borodovsky et al. 2001; Lee et al. 2016; Verma et al. 2000; Hamazaki et al. 2006; Qiu et al. 2006; Yao et al. 2006). The extrinsic ubiquitin receptors RAD23, DSK2, DDI1 may be considered to belong to this group (Elsasser and Finley 2005; Elsasser et al. 2004; Zhang et al. 2009a). Many E3 ubiquitin ligases, such as UBE3C/Hul5, parkin, and UBE3A/E6AP, are transiently associated with the proteasome (Crosas et al. 2006; Leggett et al. 2002; Sakata et al. 2003; Martinez-Noel et al. 2012; Kuhnle et al. 2018). Other E3s such as Ur1, anaphase-promoting complex/cyclosome (APC/C), SCFCDC4, UFD4 and some E2 enzymes have also been suggested to transiently associated with the proteasome (Demartino and Gillette 2007; Xie and Varshavsky 2000, 2002).
The second group includes other cellular factors that regulate proteasome functions in a non-essential, auxiliary fashion. For example, Ecm29 (Extracellular mutants 29) is a 205-kDa HEAT-repeat-containing protein that recognize a RP-CP intermediate (Leggett et al. 2002). Another example is a proline-rich protein called PI31 (Proteasomal Inhibitor of 31kD) that suppresses proteasome activities by preventing the association of RP and CP and regulates proteasome transport in axons (Zaiss et al. 1999; McCutchen-Maloney et al. 2000; Zaiss et al. 2002; Bader et al. 2011; Li et al. 2014; Minis el al. 2019). In yeast, Ecm29 appears to suppress the ATPase activity of the proteasome (Lee et al. 2011; De La Mota-Peynado et al. 2013). Ecm29 binds the RP-CP intermediate when the CP assembly is prematurely paused due to lack of specific β-subunits (Lehmann et al. 2010). The RP recruits Ecm29 in response to oxidative stress, which induces proteasome disassembly (Park et al. 2011; Wang et al. 2010). It was suggested that the free CP but not the 26S proteasome mediates degradation of oxidized proteins (Davies 2001; Breusing and Grune 2008). Ecm29-regulated disassembly of the proteasome increases the abundance of the free CP and allows cells to adapt to the oxidative stress (Haratake et al. 2016). In addition, by associating with various molecular motors and endosomal components, mammalian ECM29 promotes localization of the proteasome at the ER, the centrosome and likely other cellular locations (Gorbea et al. 2004; Gorbea et al. 2010).
Proteasome Destruction
Autophagic Control of Proteasome
Proteasomes are stable complexes exhibiting a half-life of 16 h in the embryonic fibroblasts of mouse and more than 2 weeks in the liver cells of rat (Tanaka and Ichihara 1989; Pack et al. 2014; Tomita et al. 2019). Their degradation could be also rapid under specific conditions. Three pathways have been discovered to control proteasome turnover (Marshall and Vierstra 2019; Karmon and Ben Aroya 2019). The first pathway is by caspase-catalyzed cleavage. Upon apoptosis induction, RPN2, RPN10 and RPT5 are cleaved by the caspase-3, which impairs proteasome activity in human cells (Sun et al. 2004). In Drosophila melanogaster, the caspase-3 cleaves the α2, α4, β4, and RPT1 subunits (Adrain et al. 2004). The second pathway is by the breakdown of dysfunctional proteasome subunits by the UPS itself to avoid their mis-assembly into the proteasome holoenzyme. It was shown that yeast Hsp42 coalesces the dysfunctional subunits into cytoplasmic condensates that can be removed by the proteasome itself (Peters et al. 2015; Nahar et al. 2019). The third pathway is by proteaphagy that relies on the autophagy system, which degrades large heterogeneous cytoplasmic constituents, such as organelles, protein aggregates and invasive pathogens (Marshall et al. 2015; Marshall et al. 2016; Marshall and Vierstra 2015).
There are two forms of proteaphagy, one that is induced by nutrient starvation and the other that clears inactive proteasomes through ubiquitylation of dysfunctional proteasome itself (Marshall and Vierstra 2015; Marshall et al. 2015; Marshall et al. 2016; Cohen-Kaplan et al. 2016; Waite et al. 2016; Nemec et al. 2017). During the autophagy process, cytoplasmic materials are delivered to the lysosome in mammals or the vacuole in plants and yeast for breakdown by resident hydrolases (Reggiori and Klionsky 2013; Gatica et al. 2018; Marshall and Vierstra 2018a; Levine and Kroemer 2019). During the macroautophagy process, a cup-shaped, isolated membrane named the phagophore is formed de novo and suspended in the cytoplasm. An autophagosome is eventually formed upon the closure of elongated phagophore into a double-layer membrane sphere. The outer membrane of the autophagosome then fuses with the lysosome or vacuole to form an autophagic body. By breaking the inner membrane, the cargos inside the autophagosome are exposed to the entire autophagic body and broken down by lysosomal or vacuolar hydrolases at acidic pH optima, recycling constituents of amino acids, nucleotides, carbohydrates and fatty acids for new biosynthesis (Parzych and Klionsky 2019).
The autophagy is carried out by a series of autophagy-related (ATG) proteins. Upon stimulation by upstream signals from nutrient-responsive kinases, such as Tor1/2 and Snf1, the autophagy is initiated by the serine/threonine kinase complex ATG1. Membrane delivery required by the autophagy is controlled by the transmembrane protein ATG9. Autophagosome nucleation is mediated by the phosphatidylinositol-3-phosphate (PI3P) signal that is generated by the class III phosphatidylinositol-3-kinase (PI3K) complex. Membrane extension is controlled by the ATG2-ATG18 complex at the site of PI3P labeling. Cargo recruitment is mediated by the ubiquitin-like protein ATG8 (MAP1LC3 or GABARAP in mammals) and its conjugation enzymes in a way highly analogous to ubiquitylation (Ohsumi 2001; Marshall and Vierstra 2018a; Levine and Kroemer 2019). Specifically, ATG8 is activated by the E1 enzyme ATG7, transferred to the E2 enzyme ATG3, and conjugated via an ether linkage to the lipid phosphatidylethanolamine (PE) by a hexameric E3 ligase complex ATG5-ATG12-ATG16. Membrane-bound ATG8 interacts via an ATG8-interacting motif (AIM, also named an LC3-interaction region, LIR) with autophagy adaptors or receptors that recruit the vesicular transport machinery or specific cargo (Rogov et al. 2014; Farre and Subramani 2016; Gatica et al. 2018; Marshall and Vierstra 2018a; Marshall et al. 2019; Noda et al. 2008; Noda et al. 2010; Klionsky and Schulman 2014; Maqbool et al. 2016; Rogov et al. 2017).
Autophagic Clearance of Inactive Proteasome
In plants and yeast, a proteaphagic pathway independent of ATG1 can be stimulated by genetic mutations that impair proteasome assembly, by proteasome inhibitors, such as Bortezomib and MG132, and by pathogenic effectors, such as HopM1 from Pseudomonas syringae (Marshall et al. 2015; Marshall et al. 2016; Ustun et al. 2018; Nemec et al. 2017). Proteasome inhibition stimulates ubiquitylation of the proteasome itself and causes its accumulation (Book et al. 2010; Kim et al. 2013; Marshall et al. 2015; Marshall et al. 2016). It is unclear which and how proteasome subunits are ubiquitylated. The ubiquitin receptors in the autophagy system then recognize and deliver the ubiquitylated inactive proteasomes to ATG8. In Arabidopsis, RPN10 serves as the autophagy receptor, which uses two distinct UIMs to deliver proteasomes to the autophagosome. One UIM recognizes the ubiquitylated proteasome, while the other UIM recognizes ATG8 in its UIM docking site (UDS) (Marshall et al. 2015; Marshall et al. 2019). The yeast Rpn10 lacks the second UIM for Atg8 binding and has no detectable activity in proteaphagy. Instead, the yeast receptor Cue5 uses a CUE domain to recognize the ubiquitylated proteasome and an AIM domain to recognize Atg8 (Marshall et al. 2016; Lu et al. 2014).
In yeast, aggregation of the 26S proteasome into a superstructure named peri-vacuolar insoluble protein deposit (IPOD) often precedes the ubiquitylation-dependent proteaphagy (Kaganovich et al. 2008; Marshall et al. 2016). The Hsp42 chaperone mediates the formation of IPOD by coalescing aggregated proteins (Specht et al. 2011; Malinovska et al. 2012; Miller et al. 2015). The IPODs are structurally distinct from PSGs (Marshall and Vierstra 2018b; Peters et al. 2016). They can be differentiated based on the co-localized proteins, such as the prion protein Rnq1 in IPODs or Blm10 in PSGs.
The RP-CP association has been observed to be allosterically stabilized through small-molecule inhibitor engagement at the proteolytic active sites in the CP, which appears to induce autophagic degradation of both CP and RP (Kleijnen et al. 2007; Haselbach et al. 2017). By contrast, mutations that compromise the integrity of proteasome assembly induce clearance of the affected CP or RP separately (Marshall et al. 2016). Thus, the proteaphagy can be carried out for both the proteasome holoenzyme and for the free CP or RP separately. How inhibitor-induced proteaphagy discriminates between functional and dysfunctional proteasomes is very likely linked to the conformational landscape of the proteasome regulated by inhibitor interactions (Haselbach et al. 2017). It is possible that the inhibitor-bound proteasome is strictly stabilized in the conformation of the resting state (SA) that is better recognized by the ubiquitylation machinery and Hsp42, which directs their accumulation into IPODs, whereas the functional proteasome samples a significantly more expanded energy landscape that can effectively evade proteaphagy machinery (Dong et al. 2019).
Mechanism of Substrate Selection and Ubiquitin Recognition
Degradation Signals
Most proteins targeted for proteasomal degradation are recognized through ubiquitin signals (Komander and Rape 2012), although ubiquitylation is also used for signal transduction in other cellular processes (Kulathu and Komander 2012; Yau and Rape 2016). Another essential requirement for efficient substrate degradation is an unstructured initiation region in the substrate (Prakash et al. 2004; Takeuchi et al. 2007; Zhao et al. 2010). Localization of a protein with such an initiation region to the proteasome could lead to its degradation even in the absence of ubiquitylation (Janse et al. 2004; Erales and Coffino 2014; Murakami et al. 1992). Lack of an effective unfolded initiation site protects a protein from proteasomal degradation (Fishbain et al. 2011; Heinen et al. 2011; Elsasser and Finley 2005). Variations in the length and composition of the unstructured segments affect protein half-life presumably due to the changes of binding affinity with the initial substrate-binding sites in the proteasome (Fishbain et al. 2015; van der Lee et al. 2014). The requirement of an unstructured initiation segment of a substrate has been visualized in the cryo-EM structure of the proteasome in state EB, whereas the initial ubiquitin recognition steps are observed in the structures of state EA1 and EA2 (Dong et al. 2019). Although the entire ubiquitin chains have not been visualized in the proteasome, these structural snapshots suggest an intriguing model for consecutive steps in the coordinated ubiquitin recognition and the engagement of the unfolded initiation site with the proteasome, which appears to be also coordinated with the catalytic step of deubiquitylation regulated by the ATP hydrolytic cycles.
Ubiquitin Code
Polyubiquitin chains are the predominant degradation signals in vivo (Fig. 1.14) (Komander and Rape 2012). The compactness and flexibility of the polyubiquitin chains are dependent on the lysine-linkage type and serve as a “ubiquitin code” to control selectivity and specificity of substrate targeting. Ubiquitin-ubiquitin adducts have been observed to form via all possible Lys residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63) as well as the N-terminal Met1 in cells by mass spectrometry. The ubiquitin chains can be either formed by homogeneous linkage types or by heterogeneous ones leading to diverse chain topologies including branches. The most commonly detected linkages are through Lys48, followed by Lys11 and Lys63 in yeast (Xu et al. 2009) and Lys63, Lys29, and Lys11 in human (Dammer et al. 2011). The type of chain linkage is determined by the E2 enzyme that works with the E3 ubiquitin ligases or by the class of E3 enzymes that form covalent ubiquitin intermediates (i.e., the HECT and ring-between-ring E3s). Different E2s can work with the same E3 ligase to construct polyubiquitin chains of various linkages and lengths at distinct locations on the same substrate (Ye and Rape 2009). In parallel to their polymerization, ubiquitin chains are trimmed by DUBs associated with the proteasome, or free DUBs in the cytosol (Komander et al. 2009).
The ubiquitin code and initiation region of substrates. a Structures of diubiquitin chains with different linkages. The diubiquitin chains are linked through their N and C-termini (Met1) (PDB ID: 3WXE) (Sato et al. 2015), Lys6 (PDB ID: 3ZLZ) (Hospenthal et al. 2013), Lys11 (PDB ID: 2MBQ) (Castaneda et al. 2013), Lys29 (PDB ID: 4S22) (Kristariyanto et al. 2015a), Lys33 (PDB ID: 4XYZ) (Kristariyanto et al. 2015b), Lys48 (PDB ID: 1AAR) (Cook et al. 1992). b Structures of tetraubiquitin chains with two different linkages. The PDB IDs for the Lys48- and Lys63-linked tetraubiquitin chains are 2O6V and 3HM3, respectively (Eddins et al. 2007; Datta et al. 2009). c The initiation signal recognized by the AAA-ATPase channels. The ATPase structure in the substrate-bound proteasome in state EB (PDB ID: 6MSE) is shown from the perspective of side view (left) and top view (middle) (Dong et al. 2019). The right insert shows the close-up view of the substrate interaction with the pore-1 loops, with the aromatic residues in the pore loop highlighted by the transparent sphere representations. The putative location of the substrate sidechains intercalating with these pore-loop aromatic sidechains are marked by dashed red boxes
The type of chain linkage partly dictates the conformational dynamics of the polyubiquitin signals and thus contributes to the selectivity of substrate targeting by the ubiquitin receptors. The lysine residues in ubiquitin are distributed throughout the molecular surface so that different linkages result in different geometries and flexibility of the polyubiquitin chain. The N- and C-termini are located at opposite ends of the ubiquitin fold, whereas Lys63 is located near the N terminus of ubiquitin (Datta et al. 2009). As a result, both Met1-linked and Lys63-linked polyubiquitin chains adopt extended, highly flexible conformations (Liu et al. 2015; Varadan et al. 2004; Datta et al. 2009). In contrast, Lys48 is positioned roughly halfway between N- and C-termini so that Lys48-linked chains form a more compact, less flexible structure (Eddins et al. 2007). Presumably due to higher conformational entropy of polyubiquitin chains, binding of Met1-linked linear ubiquitin chains with the proteasome is weaker than that of Lys48-linked chains (Thrower et al. 2000). Lys11-linked chains also form a compact structure that is distinct from the structure of Lys48-linked chains (Castaneda et al. 2013).
The ubiquitylated proteins can be recognized by many families of proteins that contain a ubiquitin-binding domain (UBD) (Husnjak and Dikic 2012). The UBD-containing proteins themselves can also be ubiquitylated in certain cases so that ubiquitin regulates the formation of intramolecular interactions to effect specific protein function. At least 20 families of UBDs have been identified (Husnjak and Dikic 2012). Most UBDs have a length between 30 and 120 amino acids and recognize only one or two ubiquitin molecules. The interaction surface on ubiquitin facing its receptors is a hydrophobic patch centered on conserved Ile44, Leu8, and Val70 residues. A smaller area around Phe4, Thr12, and Asn2 mediates the ubiquitin recognition as an endocytosis signal but is not essential in proteasomal degradation (Beal et al. 1996; Sloper-Mould et al. 2001). Due to flexibility of polyubiquitin conformations, chains of different linkages could be recognized by the same UBDs (Alfano et al. 2016). Post-translational modifications of ubiquitin itself have been shown to regulate their interactions with UBDs (Yau and Rape 2016). One notable modification is the phosphorylation of Ser65. Ser65-phosphorylated ubiquitin binds the proteins at the surface of damaged mitochondria and leads to the clearance of the mitochondria by an autophagic process called mitophagy (Yau and Rape 2016).
Tetraubiquitin chains linked through Lys48 are the canonical signal for proteasomal targeting (Thrower et al. 2000; Pickart 2000; Komander and Rape 2012). The proteasome-binding affinity of Lys48-linked polyubiquitin chain rapidly increases with more ubiquitin units added to the same chain until it becomes a tetraubiquitin chain, beyond which the affinity slowly increases (Thrower et al. 2000). The surfaces of every second ubiquitin molecule at positions i, i +2, i + 4, etc., in Lys48-linked chains are recognized by the proteasome (Thrower et al. 2000). A single tetraubiquitin chain is much more effective in substrate delivery to the proteasome than a single monoubiquitin (Kravtsova-Ivantsiv et al. 2009). However, multiple sites of monoubiquitylation or short polyubiquitylation with different linkages can also lead to efficient degradation both in vitro and in vivo (Hofmann and Pickart 2001; Kirkpatrick et al. 2006; Saeki et al. 2009a; Xu et al. 2009; Kim et al. 2011; Lu et al. 2015; Shabek et al. 2012; Braten et al. 2016; Martinez-Fonts and Matouschek 2016).
Lys63-linked polyubiquitin chains are the second most abundant linkages (14–40% of all ubiquitin chains) in mammalian cells (Dammer et al. 2011; Kaiser et al. 2011) and the third most common chains (16%) in yeast (Xu et al. 2009). In yeast, Lys63-linked chains have been shown to mediate partial degradation of the transcription factor Mga2-p120 (Saeki et al. 2009a). Lys11-linked ubiquitin chains are about 2–5% of all ubiquitin chains in mammalian cells (Dammer et al. 2011; Kaiser et al. 2011), and are as abundant (28%) as Lys48-linked chains in yeast (Xu et al. 2009). They play a crucial role in cell cycle regulation. The E3 ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C) assembles both Lys48- and Lys11-linked ubiquitin chains on many cell-cycle regulators to control their degradation, such as Aurora A, cyclin A, cyclin B1, securin, geminin, Nek2A, Plk1, and E2F1 (Craney and Rape 2013; Peters 2006). On some substrates, APC/C first coordinates with the UBE2C to synthesize short chains with Lys48, Lys63 and Lys11 linkages (Jin et al. 2008). The chains could be further extended by APC/C working with the UBE2S that primarily synthesizes Lys11-linked chains (Williamson et al. 2009; Wu et al. 2010). Interestingly, APC/C-synthesized branched polyubiquitin chains containing both Lys11 and Lys48 linkages are more efficient degradation signals than homogeneous Lys11-linked chains (Meyer and Rape 2014). Lys11-linked chains mediate proteasomal degradation of the hypoxia-inducible factor 1α (HIF-1α) (Bremm et al. 2014), as well as targeted degradation in response to the endoplasmic-reticulum stress (Xu et al. 2009). IRF3 degradation can be targeted by both Lys11- and Lys48-linked chains, which is induced by interferon pathways after viral infection (Qin et al. 2014). In Drosophila, the E3 ligase Cul1-Slimb synthesizes similar portions of ubiquitin conjugates with Lys11 and Lys48 linkages on the transcription factor Cubitus interruptus (Ci) targeted for partial degradation by the proteasome (Zhang et al. 2013).
Tandem repeats of the ubiquitin sequence are encoded in the UBI4 gene in yeast, and in the UBB and UBC genes in human genome. They express Met1-linked linear polyubiquitin chains that can be cleaved into individual ubiquitin molecules by ubiquitin C-terminal hydrolases. Linear ubiquitin chains can also be synthesized by the 600-kDa linear ubiquitin chain assembly complex (LUBAC) and are used as a scaffold in the assembly of signaling complexes in several pathways (e.g., NF-κB, ERK, Wnt, TNF) (Kirisako et al. 2006; Iwai et al. 2014; Shimizu et al. 2015). The linear polyubiquitin chains do not mediate proteasomal degradation in vitro (Martinez-Fonts and Matouschek 2016). However, a linear chain with a minimum of tetraubiquitin is sufficient for proteasomal targeting assisted by the Cdc48-Npl4-Ufd1 complex in vivo (Kirisako et al. 2006; Zhao and Ulrich 2010), and physiologically mediate the degradation of protein kinase C and TRIM25 (Inn et al. 2011; Nakamura et al. 2006).
The ubiquitin code is not exclusively used by the proteasome but also involved in other proteasome-independent processes such as innate immunity and chromatin remodeling (Komander and Rape 2012; Kulathu and Komander 2012). In yeast, Met4 (Flick et al. 2004) and Cdc34 (Fishbain et al. 2015; Goebl et al. 1994) can be ubiquitylated with Lys48-linked polyubiquitin chains that do not lead to proteasomal degradation. Shorter ubiquitin chains or Lys63-linked polyubiquitin chains are involved in processes like intracellular membrane trafficking during autophagy and endocytosis (Erpapazoglou et al. 2014), DNA repair (Schwertman et al. 2016), cell signaling (Chen and Sun 2009), immune responses (Wu and Karin 2015), mRNA splicing (Song et al. 2010), and the regulation of translation (Silva et al. 2015; Spence et al. 2000). Lys11-linked polyubiquitin chains target MHC-I receptor (Boname et al. 2010) for internalization from the cell surface by endocytosis. Heterotypic linkages mixed with Lys11 and Lys63 on receptor-interacting serine/threonine-protein kinase 1 (RIP1) mediate its binding to the IκB kinase (IKK) complex when the transcription factor NF-κB is activated by TNF-α (Dynek et al. 2010). Monoubiquitylation also functions in membrane trafficking as an endocytosis signal, in the regulation of transcription and in the DNA damage response (Hurley and Stenmark 2011; Ramanathan and Ye 2012).
Initiation Region of Substrates
The presence of an unfolded initiation region is an indispensable prerequisite for initiating substrate degradation by the proteasome (Kraut and Matouschek 2011; Prakash et al. 2009; Yu and Matouschek 2017). Such an unstructured region was found to either flank at the terminus or appear as an internal loop of the substrate. The receptor sites for this initiation region in the proteasome is expected to be the interior of the axial channel of the AAA-ATPase motor in the base (Fig. 1.14c) (Dong et al. 2019). As illustrated in the atomic structure of substrate-bound proteasome in state EB representing the initial commitment step (Dong et al. 2019), the minimum length requirement for a terminal initiation region spans from the ubiquitin-linking isopeptide bond, across the OB-ring, to the inner end of the ATPase channel, which is equivalent to ∼20–30 amino acids (Fig. 1.14c). This structural estimation is well consistent with several biochemical studies (Prakash et al. 2004; Takeuchi et al. 2007; Verhoef et al. 2009; Yu and Matouschek 2017). The distance of the initiation region from the substrate-conjugated ubiquitin signal apparently determines the initial affinity of the substrate binding to the AAA channel and hence influences the rate of proteasomal processing (Inobe et al. 2011; Lu et al. 2015; Fishbain et al. 2011). Proteasome-associated proteins such as extrinsic UBL-UBA receptors lacking flexible segments suitable for initial engagement can thus avoid proteasomal degradation. For instance, an unstructured segment intrinsic to RAD23 is not long enough to mediate its recognition by the proteasomal ATPase motor, whereas artificial introduction of a flexible initiation loop at the terminus of RAD23 leads to its proteasomal degradation (Fishbain et al. 2011).
An unstructured initiation loop with a sufficient length is necessary for committing substrates to the proteasome for efficient deubiquitylation and translocation (Peth et al. 2010). This commitment step is crucial for coupling of substrate deubiquitylation with translocation at the proteasome, as shown in the state-EB structure of the proteasome (Fig. 1.14c) (Dong et al. 2019; Worden et al. 2017). Decoupling of substrate translocation with deubiquitylation can reverse the commitment step, leading to futile substrate processing by the proteasome. The sequence composition of the initiation region has strong correlative effects on the rate of substrate degradation both in vivo and in vitro (Fishbain et al. 2015; Kraut et al. 2012; Yu et al. 2016b). Compared with diverse initiation sequences, those with small sidechains or lower structural complexity (i.e., stretches of glycines, alanines or serines) impair substrate degradation (Hoyt et al. 2006; Sharipo et al. 1998; Yu et al. 2016b). Chemical properties of the initiation region, such as hydrophobicity, electrostatic charge and flexibility further modulate its recognition by the proteasome.
The sequence dependence of the initiation region in the substrate could be explained at least in part by its interaction with the pore loops in the AAA domains of the proteasomal ATPase ring (Fig. 1.14c). In the state-EB structure of the proteasome, the aromatic residues in the pore-1 loops appear to touch the substrate mainchain backbone and occupy the space between adjacent side chains of the fully stretched substrate polypeptide that assumes a conformation similar to a β-strand. The staircase of the pore-loop aromatic residues intercalating substrate sidechain resembles a “rachet wheel” gripping a “cable tie” via its “teeth”. The force generated through this “ratchet teething” model will be dependent of the depth of the “rachet teeth” of the substrate. Large sidechains on the substrate polypeptide chain allow a tighter grip, which can both prevent backsliding and apply a high enough unfolding force. Indeed, introducing a low-complexity sequence of small sidechain residues at the terminus of a highly stable globule domain appears to compromise the gripping force generated by the ATPase motor, thus promoting substrate slippage and releasing partially processed substrate (Hoyt et al. 2006; Sharipo et al. 1998; Kraut and Matouschek 2011; Kraut et al. 2012; Yu et al. 2016b; Tian et al. 2005). Thus, an unstructured but slippery internal loop can serve as degradation-stop signals and allow for partial substrate degradation by the proteasome that then activates transcription factors such as NF-κB, Spt23, and Mga2 (Tian et al. 2005; Piwko and Jentsch 2006; Hoppe et al. 2000, 2001).
Ubiquitin Recognition at the Proteasome
The recruitment of ubiquitylated substrates to the proteasome can be mediated through direct ubiquitin binding with the intrinsic receptors RPN1, RPN10 and RPN13. These intrinsic receptors can also indirectly recruit ubiquitylated substrates by reversibly binding the UBL domains of extrinsic ubiquitin receptors, also known as shuttle factors, that are canonically made of UBL-UBA proteins, such as RAD23, DSK2, and DDI1 families. In yeast, the cellular abundances of the extrinsic and intrinsic receptors are quite comparable (Ghaemmaghami et al. 2003). The intrinsic ubiquitin receptors also provide docking sites for certain DUBs and E3/E4 ligases, which builds up additional layers of complexity in the functional regulation of the proteasome. Binding of either UBL-containing proteins or ubiquitin chains alone, mostly via the intrinsic ubiquitin receptors, are found to generally stimulate the ATPase activity and peptide hydrolysis in the proteasome (Collins and Goldberg 2020; Ding et al. 2019; Kim and Goldberg 2018). This effect suggest that, similar to the influence of substrate engagement, ubiquitin or UBL binding re-equilibrates the ensemble of conformational states of the proteasome toward the ED-like states (Table 1.2 and Fig. 1.9) (de la Pena et al. 2018; Ding et al. 2019; Dong et al. 2019).
RPN1
The importance of RPN1 is reflected in its ability to recognize both polyubiquitin chains and UBL domains of UBL-UBA proteins. RPN1 is structurally homologous to RPN2, which anchors another ubiquitin receptor RPN13. Both RPN1 and RPN2 resemble a tobacco pipe, with a central proteasome/cyclosome (PC) domain of 11 leucine-rich repeats (LRR) that make up the toroid-shaped “barrel” of the pipe (Fig. 1.15). This toroidal “barrel”, composed of only tightly packed α-helices, is demarcated on one side by an N-terminal α-helical domain comprising the wedge-like stem, and on the orthogonal side by a C-terminal domain (CTD) comprising mostly β-stranded barrel-like globular fold (He et al. 2012; Dong et al. 2019). In the toroidal PC domain of RPN1/RPN2, the pore formed by the LRRs is occupied by two α-helices. This packing is mediated by the highly hydrophobic residues in these two central α-helices that sequester them from solvent.
Long-range interaction between RPN1 and RPN2 (PDB IDs: 6MSJ, 6MSK) (Dong et al. 2019). a Long-range association between RPN1 and RPN2 via a long looping (residue 820–871) structure extending from the RPN2 PC domain observed in state ED1. b Comparison of the RPN1-RPN2 structures between states ED1 and ED2 shows a prominent translation of RPN1 relative to the CP
RPN1 and RPN2 are positioned distally from the proteasomal long axis and are the two most surface-exposed RP subunits. RPN1 and RPN2 engage with the RPT1-RPT2 and RPT3-RPT6 CC domains, respectively. In RPN2, both the N-terminal wedge-like domain and the concave side of the PC domain bind the N-terminal CC of RPT3 and RPT6, with the PC domain and CTD being positioned above the entrance of the OB ring. Similarly, the N-terminal CC domains of RPT1 and RPT2 interact with the concave side of the PC domain of RPN1 (Fig. 1.15). The RPN1 PC domain and the RPT1-RPT2 CC domain form a surface cavity that is inserted by a short α-helix from RPN2. This α-helix is the middle part of a long loop (residues 820–871) extending from the RPN2 PC domain. Through such a long-range interaction of RPN1-RPN2, a more extensive interface is stabilized between RPT1-RPT2 and RPN1-RPN2 at the concave side of the RPN1 PC domain facing RPN2. Such a quaternary interface is best reconstructed in states ED1 and ED2, but less clearly observed in other states (EA-C) (Dong et al. 2019). It appears that RPN1 is most dynamic in state EC, presumably due to the requirement of ubiquitin release following deubiquitylation.
Structural and mutagenesis studies have revealed that the yeast Rpn1 harbors two receptor sites, designated T1 and T2, which preferentially bind ubiquitin and UBL proteins, respectively (Fig. 1.16) (Shi et al. 2016; Chen et al. 2016b). Although the T2 site was found to only bind UBL proteins such as RAD23, cryo-EM structures of the substrate-bound human proteasome suggest that this site can also engage polyubiquitin (Dong et al. 2019). It is likely that the ubiquitin or UBL-binding affinity is different between the two sites, making one site primary and the other auxiliary under certain scenarios. When the primary site captures a polyubiquitin chain, the auxiliary site could still directly bind a distal ubiquitin molecule in the chain despite lower local affinity.
Mechanism of ubiquitin recognition by the 26S proteasome (PDB IDs: 6MSB, 6MSD, 6MSE) (Dong et al. 2019). a The cryo-EM maps of states EA1 and EA2. b Ubiquitin densities in state EA1 (left) and EA2 (right). The T1 and T2 sites are labelled by fitting the yellow cartoon representation of the NMR structure (PDB ID 2N3U) of RPN1 T1/T2 element in complex with two ubiquitin molecules into our density, showing the ubiquitin on RPN1 is bound to a site very close to the T1/T2 sites (Shi et al. 2016). The density maps are low-pass filtered to 8 Å to show the ubiquitin features clearly, due to the lower local resolution of the ubiquitin density in these maps. c Comparison of two ubiquitin moieties between RPN11 and RPT4/5 CC among EA1, EA2 and EB. The cryo-EM densities rendered as grey mesh representations are low-pass filtered to 8 Å. The atomic model of ubiquitin is shown as a magenta cartoon representation
RPN10
RPN10/S5a seems to represent a “canonical” intrinsic ubiquitin receptor in the proteasome, given its unique proximity to the essential DUB RPN11 than any other intrinsic ubiquitin receptors in the proteasome. RPN10 recognizes polyubiquitin chains via its α-helical UIM domains (Deveraux et al. 1994; Fu et al. 1998; van Nocker et al. 1996). RPN10 includes one to three UIMs at its C terminal region. The exact number of UIMs in RPN10 depends on the species. Yeast Rpn10 that possesses only a single UIM still prefers polyubiquitin chains over monoubiquitin (Zhang et al. 2009a, d). Solution NMR structural studies indicate that the two UIMs of the human RPN10 greatly enhance its affinity for a Lys48-linked ubiquitin chain (Zhang et al. 2009d). Although not observed in the context of the proteasome, there is a preference for UIM1 on recognizing the distal ubiquitin and for UIM2 binding the proximal one; when both RPN10 and RPN13 bind the same Lys48-linked polyubiquitin chain, RPN13 seems to recognize the distal ubiquitin whereas the UIMs compete for the proximal ones (Zhang et al. 2009d). This observation is consistent with the spatial arrangement of RPN10 being much closer to the DUB RPN11 and OB ring than RPN13 in the proteasome.
The UIMs are highly dynamic and appear as a poorly resolved diffuse density in cryo-EM structures of the proteasome (Dong et al. 2019). In contrast, the N-terminal VWA domain of RPN10 is tightly folded and well resolved in the cryo-EM structures (Dong et al. 2019; Riedinger et al. 2010). The VWA domain makes extensive contact with the lid subunits RPN8 and RPN9, suggesting that it structurally stabilizes the quaternary architecture of the lid and thus of the RP. Disruption of the RPN10 VWA domain or loss of RPN10 destabilizes the lid-base interaction (Glickman et al. 1998; Fu et al. 2001; Tomko and Hochstrasser 2011). RPN10 also provides primary binding sites to recruit extrinsic ubiquitin receptors like DSK2/Ubiquilin (Matiuhin et al. 2008; Zhang et al. 2009a; Chen et al. 2019) and certain E3 ligases like E6AP/UBE3A (Buel et al. 2020). Binding of the E6AP AZUL (Amino-terminal Zinc-binding domain of Ubiquitin E3a Ligase) domain induces refolding of an unstructured C-terminal segment in RPN10 into a helical bundle (Buel et al. 2020). The yeast Rpn10 is monoubiquitylated in vivo, at Lys71, Lys84 and Lys99 in the VWA domain, which regulates its interactions with substrates by inhibiting the UIM (Isasa et al. 2010). The levels of Rpn10 monoubiquitylation are reversibly controlled by Rsp5, an E3 in the NEDD4 ubiquitin ligase family, and a DUB Ubp2. Interestingly, cold shock, heat shock and cadmium all seem to reduce Rpn10 monoubiquitylation, suggesting that this modification is likely a mechanism of proteasome regulation in response to proteotoxic stress (Isasa et al. 2010).
Although the RPN10 UIMs are poorly resolved in the cryo-EM reconstructions of the human proteasome, there appears to be multiple ubiquitin densities in states EA1, EA2 and EB near RPN10 (Fig. 1.16). The low-resolution density of RPN10 UIMs in state EA1 appears to connect with the N-terminal helices of RPT4-RPT5 CC presumably via the polyubiquitin chains bound to the UIMs. Two ubiquitin molecules likely connected within the same chain are attached to the near and far sides of RPT4-RPT5 CC immediately next to the RPN10 UIMs (Dong et al. 2019; Chen et al. 2020). Consistently, in state EA2, one ubiquitin is transferred to the ubiquitin-binding site of RPN11, whereas the density between RPN10 UIMs and RPT4-PRT5 CC appears to be broken (Fig. 1.16b). As the step of substrate deubiquitylation is presumably represented by state EB, these structural observations suggest that multiple steps of ubiquitin transfer and repositioning or remodeling of substrate-conjugated polyubiquitin chains are required for optimal deubiquitylation (Dong et al. 2019). A flexible link of UIMs to the VWA domain of RPN10 would permit conformational adaptability of the receptor sites in recognizing and delivering polyubiquitin chains of diverse geometries to the deubiquitylation site. In this context, the RPT4-RPT5 CC provides auxiliary receptor sites for enhanced docking of RPN10 UIM-recognized ubiquitin chains and assisting peptide-proximal ubiquitin transfer to the DUB RPN11 for efficient substrate deubiquitylation (Dong et al. 2019; Lam et al. 2002).
RPN13
RPN13 prefers the recognition of the canonical Lys48-linked ubiquitin signals. The N-terminal region of RPN13 comprises the Pleckstrin-like Receptor for Ubiquitin (PRU) domain that structurally resembles a pleckstrin homology fold (Schreiner et al. 2008; Husnjak et al. 2008). The PRU domain of RPN13 selectively recognizes a dynamic, extended conformation of Lys48-linked diubiquitin via loops extending from the β-strands in the PRU domain and exhibits ∼90 nM KD for Lys48-linked diubiquitin. In the proteasome, the PRU domain binds a flexible C-terminal region of RPN2, which emanates from its LRR-like toroidal domain (Chen et al. 2010; Sakata et al. 2012; Lu et al. 2017a, 2020). The recruitment of RPN13 near the toroid apex of RPN2 positions its PRU domain about four-ubiquitin away from the entrance of the ATPase ring, which is comparable to the distance from the ubiquitin-binding sites in RPN1 and RPN10 (Lu et al. 2020). The PRU domain of human RPN13 is connected by a flexible linker to a C-terminal helical DEUBAD (DEUBiquitinase ADaptor) domain that recruits and activates the DUB UCH37 (Lu et al. 2017a; Sahtoe et al. 2015; Hamazaki et al. 2006; Qiu et al. 2006; Yao et al. 2006). In contrast to the occupancy of the intrinsic receptors typically at more than 50% in the yeast proteasome (Sakata et al. 2012), RPN13 seems to be either substoichiometric or flexibly bound in the endogenously purified human proteasome (Dong et al. 2019).
NMR studies have shown that in the absence of the proteasome, multiple binding modes are feasible for RPN10 and RPN13 in recognition of Lys48-linked tetraubiquitin chains (Zhang et al. 2009d). This prompts the possibility of promiscuous modes of polyubiquitin chain recognition by the intrinsic ubiquitin receptors in the context of the proteasome. It is conceivable that RPN1, RPN10 and RPN13 simultaneously bind polyubiquitin chains to orient substrates for optimal engagement and deubiquitylation (Zhang et al. 2009d; Liu et al. 2019; Lu et al. 2020). This multi-site recognition is both kinetically and enthalpically favored and could be functionally adapted to tackle the vast conformational diversity of the polyubiquitin chains conjugated on protein substrates. Although substrates with multiple short ubiquitin chains can be targeted by any of the known receptors for proteasomal degradation, those delivered to the proteasome via a UBL domain were observed to be degraded more efficiently by RPN13 and RPN1 (Martinez-Fonts et al. 2020). It remains to be clarified if RPN10 and RPN13 can partly function as a substrate shuttle like those extrinsic receptors to some extent.
Delivery of Substrates to the Proteasome
Shuttle Proteins
The extrinsic ubiquitin receptors share common domain organizations. The N-terminal UBL domain that can be recognized by an intrinsic ubiquitin receptor in the proteasome is integrated in a single fold with C-terminal UBA domains that recognize ubiquitin signals (Elsasser et al. 2002; Funakoshi et al. 2002; Schauber et al. 1998; Kaplun et al. 2005; Hofmann and Bucher 1996). Whereas Saccharomyces cerevisiae contains one shuttle receptor in each family of Rad23, Dsk2 and Ddi1, higher eukaryotes contain many more paralogs, such as RAD23A/hHR23A, RAD23B/hHR23B, UBQLN1/2/3/4 and DDI1/2, and other additional UBL-UBA proteins, such as UBAC1, UBL7 and NUB1L. Different paralogs of shuttle-receptor have distinct binding preferences for specific intrinsic ubiquitin receptors owing to differences in their individual UBL interfaces (Chen et al. 2016b). The associations between intrinsic and extrinsic ubiquitin receptors are highly dynamic and complicate their structural analysis. Some UBL-containing proteins can support proteasome delivery in vitro but fail in vivo (Yu et al. 2016a; Gomez et al. 2011; Walters et al. 2002). Differences in the number of UIMs of RPN10 create another level of complexity in the preferential proteasomal binding sites for the UBL domains of the extrinsic receptors. The N-terminal UIM1 of human RPN10 exhibits 25-fold stronger affinity over UIM2 for binding the UBL domain of UBQLN2 (Chen et al. 2019). Recent NMR structural studies on RAD23 and UBQLN2/hPLIC2 examined the structural basis for their preferences to bind the RPN1 and RPN13, respectively (Fig. 1.17) (Chen et al. 2016b; Shi et al. 2016; Chen et al. 2019).
The architecture of ubiquitin receptors and their interactions with ubiquitin. a Domain organization of the ubiquitin receptors RPN10, RPN13, RAD23, DSK2 and DDI1. b Distinct modes of interaction with ubiquitin are shown for each receptor. Amino acids of each ubiquitin-binding domain that contact ubiquitin are highlighted. The structures correspond to PDB IDs as follows: Ly48-linked diubiquitin-bound yeast RPN1 toroid segment (2N3V) (Shi et al. 2016), Lys48-linked diubiquitin-bound human RPN10 UIM (2KDE) (Zhang et al. 2009d), Lys48-linked diubiquitin-bound human RPN13 PRU (5YMY) (Liu et al. 2019), Lys48-diubiquitin-bound human RAD23A/hHR23A UBA (1ZO6) (Varadan et al. 2005), and ubiquitin-bound yeast Dsk2 UBA (1WR1) (Ohno et al. 2005). PRU, Pleckstrin-like receptor for ubiquitin domain; UBA, ubiquitin-associated domain; UBL, ubiquitin-like domain; UIM, ubiquitin-interacting motif; VWA, von Willebrand factor A domain; UBA, ubiquitin-associated domain
Besides the ubiquitin receptors in the UBL-UBA family, other proteins not in this family have also been found to function as a shuttle factor (Saeki 2017; Finley and Prado 2019). Several members in the ZFAND family of zinc-finger proteins, including ZFAND1, ZFAND2A/B and ZFAND5, are among such candidates (Stanhill et al. 2006; Yun et al. 2008; Osorio et al. 2016; Rahighi et al. 2016; Turakhiya et al. 2018; Hishiya et al. 2006; Lee et al. 2018). Recruiting extrinsic ubiquitin receptors with a flexible domain architecture may allow the proteasome to accommodate a greater variety of ubiquitin signals and substrate structures than those readily recognized by the intrinsic ubiquitin receptors alone. They can also offer substrate selectivity and specificity upstream of ubiquitylation by binding disordered regions in substrates. Using their UBL domains to bind both E4 ubiquitin ligases and intrinsic ubiquitin receptors allows them to ferry ubiquitylated proteins into the proteasome for degradation (Elsasser et al. 2004; Chen and Madura 2002; Kim et al. 2004; Hanzelmann et al. 2010; Tsuchiya et al. 2017). As a result, mistargeted or misfolded proteins that can not be directly recognized by the proteasome due to lack of ubiquitin chains or deficiency of an unstructured initiation region can still be delivered to the proteasome for breakdown (Hanzelmann et al. 2010).
RAD23
RAD23 contains four domains: UBL, UBA1, STI1/XPCB and UBA2 (Walters et al. 2003). The UBA1 and C-terminal UBA2 are separated by the STI1/XPCB (heat-shock chaperone-binding) domain (Yokoi and Hanaoka 2017). UBA1 and UBA2 preferentially bind Lys63-linked and Lys48-linked polyubiquitin chains, respectively, although both can recognize Lys48-linked chains (Raasi et al. 2005). RAD23 UBL domain binds the T1 and T2 sites on the toroidal domain of RPN1 (Elsasser et al. 2002; Shi et al. 2016), positioning it near the entrance of the central channel of the base leading to the CP gate (Rosenzweig et al. 2008). RAD23 may also interact with RPN10 and presumably other ubiquitin receptors via its UBL domain (Ishii et al. 2006). Phosphorylation of its UBL domain inhibits RAD23 interaction with the ubiquitin receptors in the proteasome (Liang et al. 2014). The RAD23 UBL domain can also bind other proteins that can recognize ubiquitin-like structures. For example, it is recognized by the yeast E4 enzyme Ufd2 to facilitate proteasomal degradation of ubiquitin-fusion degradation (UFD) substrates (Kim et al. 2004; Hanzelmann et al. 2010). The peptidyl tRNA hydrolase Pth2 binds the RAD23 UBL, which antagonizes ubiquitin-dependent proteolysis by inhibiting the RAD23 association with the proteasome (Ishii et al. 2006).
The roles of RAD23 in degradation appear to be adaptive and dynamically regulated. Several studies suggest that it suppresses proteasomal degradation under certain conditions (Ortolan et al. 2000; Raasi and Pickart 2003), whereas others have found that it promotes degradation by shuttling protein substrates to the proteasome (Shi et al. 2016; Verma et al. 2004; Elsasser et al. 2004). RAD23 delivers substrates preprocessed by p97/Cdc48 (Olszewski et al. 2019) to the proteasome by protecting the substrates from further processing of their ubiquitin chains, including chain elongation by E4 ligases and deubiquitylation by cytosolic DUBs (Tsuchiya et al. 2017). This is expected to efficiently escort the substrates to the proteasome by preventing premature processing of the ubiquitin signals (Richly et al. 2005). Interestingly, RAD23 is found to be necessary for the formation of nuclear proteasome foci, the transient structures that contain p97/VCP and multiple proteasome-interacting proteins and collectively constitute a proteolytic entre in the nucleus (Yasuda et al. 2020). A liquid-liquid phase separation can be triggered by multivalent interactions of two UBA domains of RAD23 and polyubiquitin chains (Yasuda et al. 2020). RAD23 also functions in the pathway of endoplasmic reticulum-associated degradation (ERAD) by binding of its Rad4-binding STI1/XPCB domain to the deglycosylase Png1, forming a complex that mediates proteasomal degradation of a subset of glycosylated ERAD substrates (Kim et al. 2006). Knockdown of RAD23 genes in mouse models and human cells demonstrated their importance in mammalian development, cell cycle control and apoptosis (Yokoi and Hanaoka 2017).
Ubiquilins/Dsk2
Ubiquilins, the mammalian orthologs of the yeast Dsk2, are a conserved family of four ubiquitin-like proteins (ubiquilin-1-4 or UBQLN1-4) that function as shuttle proteins and deliver ubiquitylated substrates to the proteasome. Similar to RAD23, ubiquilins harbor an N-terminal UBL domain that binds the intrinsic ubiquitin receptors in the proteasome and a C-terminal UBA domain that recruits a substrate (Ko et al. 2004; Hamazaki et al. 2015). Mutations in ubiquilins that compromise their ability to bind the intrinsic ubiquitin receptors were linked to elevated cellular levels of ubiquitylated proteins and aggregate formation, which are implicated in the pathogenesis of several neurodegenerative diseases, such as Huntington’s and Alzheimer’s diseases, and amyotrophic lateral sclerosis (ALS) (Wang and Monteiro 2007; Wang et al. 2006; Haapasalo et al. 2011; Hjerpe et al. 2016). In yeast, overexpression of Dsk2 impairs proteolysis and exerts a cytotoxic effect (Funakoshi et al. 2002; Biggins et al. 1996; Matiuhin et al. 2008). This effect seems to be attenuated by binding of the Dsk2 UBL to the extraproteasomal Rpn10 UIM that restricts Dsk2 access to the proteasome and alleviates the cellular stress associated with Dsk2 (Matiuhin et al. 2008). This interaction may be regulated by Rpn10 monoubiquitylation (Zuin et al. 2015), which primarily regulates Rpn10 UIM binding with polyubiquitylated substrates (Isasa et al. 2010; Woelk et al. 2006; Di Fiore et al. 2003).
Ubiquilins have been demonstrated to mediate the degradation of damaged proteins in response to oxidative stress (Liu et al. 2014). Recent studies have revealed a pathway for maintaining protein homeostasis mediated by UBQLN2 (Hjerpe et al. 2016) and a similar role of yeast Dsk2 (Samant et al. 2018) in the breakdown of misfolded or damaged proteins in the nucleus generated by heat shock. The UBQLN2 protein is found to cooperate with HSP70-HSP10 disaggregase chaperone machinery to clear protein aggregates, such as polyglutamine-expanded Huntingtin, via the proteasome (Hjerpe et al. 2016). UBQLN2 recognizes substrate-bound HSP70 and delivers it to the proteasome to enable the degradation of damaged, misfolded or aggregated proteins in the nucleus. The absence of autophagy in the nucleus underscores the importance of this substrate-targeting pathway to the nuclear proteasome. Further studies of similar pathways in yeast suggest that nuclear Dsk2 shuttles Lys48-linked polyubiquitylated substrates to the nuclear proteasome, whereas in the cytoplasm, hybrid Lys11/Lys48 chains enhance the affinity of misfolded proteins by the cytoplasmic proteasome, presumably by engaging multiple ubiquitin receptors at the same time (Samant et al. 2018).
P97/Cdc48
In addition to the extrinsic ubiquitin receptors, other proteins may also participate in shuttling of substrates to the proteasome. The double-ring-shaped hexameric AAA+ ATPase unfoldase p97 or valosin-containing protein (VCP) in mammalian cells, and its yeast ortholog Cdc48, remodel or segregate ubiquitylated substrates for ubiquitin-dependent degradation (Stolz et al. 2011; Xia et al. 2016; van den Boom and Meyer 2018; Meyer et al. 2012). They play an important role in the UPS and protein quality control (Godderz et al. 2015), especially in ERAD (Wolf and Stolz 2012; Christianson and Ye 2014; Wu and Rapoport 2018; Stein et al. 2014) and outer mitochondrial membrane associated degradation (OMMAD) (Taylor and Rutter 2011; Heo et al. 2010; Xu et al. 2011). Although archaeal Cdc48 can assemble with the CP in vitro through artificial crosslinking (Barthelme et al. 2014), similar assembly has not been observed in eukaryotic cells. Since the proteasome requires an unstructured segment in its substrate to initiate substrate processing (Prakash et al. 2004; Fishbain et al. 2011; Inobe et al. 2011), p97/Cdc48 can act upstream of the proteasome to preprocess those substrates that are well-folded without a flexible initiation region or located in membranes (Beskow et al. 2009; Olszewski et al. 2019). p97/Cdc48 can partially unfold the substrate to create an unstructured initiation loop and transfer it to the UBL-UBA proteins like RAD23 and DSK2 that ultimately deliver the substrate to the proteasome for degradation (Itakura et al. 2016; Baek et al. 2011; Richly et al. 2005). Alternatively, p97/Cdc48 might completely unfold the substrate that is directly translocated into the free CP for breakdown (Barthelme and Sauer 2012, 2013). Furthermore, p97/Cdc48 is also involved in other cellular processes, such as ribosomal quality control (Verma et al. 2013; Brandman et al. 2012), extraction of chromatin-bound proteins (Ramadan et al. 2007; Franz et al. 2016; Dantuma and Hoppe 2012), membrane fusion and vesicular trafficking (Bug and Meyer 2012; Patel et al. 1998), and autophagy (Ju et al. 2009; Papadopoulos et al. 2017).
The p97/Cdc48 complex functions as a molecular motor that mechanically unfolds a substrate translocating across its axial channel. Each p97/Cdc48 protomer comprises two tandem ATPase domains (D1 and D2) that are homologous to the proteasomal RPT subunits, and couple ATP hydrolysis to its pore-1/2 loops that can translocate substrates across the axial channel of the p97/Cdc4 homohexamer (Sauer and Baker 2011; Ogura and Wilkinson 2001; Lupas and Martin 2002; Iyer et al. 2004; Hanson and Whiteheart 2005; Erzberger and Berger 2006; Pamnani et al. 1997). The N domains are not fixed with respect to the double ring (Zhang et al. 2000; Davies et al. 2008). Upon ATP binding to the D1 ATPases, the N domains change from a “down conformation” coplanar with the D1 ring to an “up conformation” above the D1 plane (Banerjee et al. 2016; Tang et al. 2010). To engage a substrate, p97/Cdc48 uses other cofactors such as the Ufd1-Npl4 (UN) heterodimer, which binds the N domains and acts as the ubiquitin receptor (Hanzelmann and Schindelin 2017; Buchberger et al. 2015). To achieve optimal substrate binding, at least five ubiquitin moieties are required in a ubiquitin chain on a substrate (Bodnar et al. 2018; Bodnar and Rapoport 2017). These cofactors endow p97/Cdc48 with the substrate selectivity.
Cryo-EM structures of the yeast Cdc48 in a complex with UN and a ubiquitylated substrate establish the molecular basis for its substrate processing (Cooney et al. 2019; Twomey et al. 2019). Interestingly, a ubiquitin molecule is partially unfolded and the unfolded segment binds a conserved groove of Npl4 leading to the axial channel of the double ATPase rings (Twomey et al. 2019). It is still unclear how the Cdc48-UN unfoldase complex initiates its substrate processing. But the current hypothesis is that the substrate engagement starts on a segment of the ubiquitin molecule proximal to the substrate that can subsequently lead to substrate unfolding. The D1 domains instead of the D2 are suggested to drive the translocation of ubiquitin-substrate adducts after ATP-independent insertion of the ubiquitin N-terminal segment into the axial channel (Bodnar and Rapoport 2017; Olszewski et al. 2019; Twomey et al. 2019). Importantly, this model explains why p97/Cdc48 does not need an unstructured initiation region in the substrate to start on substrate processing.
Proteasome-Associated Ubiquitin Ligases
The proteasome exhibits not only deubiquitylating activity, but in certain condition is also associated with ubiquitin ligase activity. Many ubiquitin ligases have been found to be potentially complexed with the proteasome in a transient fashion, which can impact how a substrate is recognized and processed by the proteasome (Verma et al. 2000; Xie and Varshavsky 2000, 2002; Crosas et al. 2006; Leggett et al. 2002; Besche et al. 2009; Martinez-Noel et al. 2012; Kuhnle et al. 2018). One hypothesis is that there is a mutual regulation between the proteasome and its associated ubiquitin ligases. The ubiquitylation activity of a ligase could be enhanced once it associates with both a substrate and the proteasome, as the temporal delay between ubiquitylation and deubiquitylation at the proteasome is reduced (Xie and Varshavsky 2000). Ubiquitin-conjugating activity of the yeast Hul5 ligase was proposed to act specifically on proteasome-bound substrates that have already been ubiquitylated by another E3 ligase. Essentially, Hul5 functions as a ubiquitin chain-elongation E4 enzyme regulating the proteasome activity (Crosas et al. 2006). In vitro, Hul5 does not take unmodified cyclin B as a substrate and efficiently adds ubiquitin to cyclin B only if it had already been ubiquitylated (Crosas et al. 2006).
Mechanism of Substrate Deubiquitylation
Once ubiquitin receptors deliver a polyubiquitylated substrate to the proteasome, the ubiquitin signal must be removed from the substrate by the DUBs in the proteasome to enable transport of the unfolded substrate to the proteolytic sites in the CP chamber. Although there are about a hundred distinct DUBs encoded in the human genome, which govern a vast diversity of ubiquitylation pathways (Leznicki and Kulathu 2017), only a few DUBs were found to physically associate with the proteasome. By recycling ubiquitin from substrates, DUBs positively regulate the cellular level of free ubiquitin available for ubiquitylation and, by dynamically reversing ubiquitylation, decide the fate of protein substrates in myriad cellar processes. The isopeptidase activity of proteasome-associated DUBs is often quite low in their isolated form and is prominently enhanced upon their incorporation into the proteasome. The allosteric regulation is reciprocal or bidirectional between the DUBs and the proteasome, since multiple catalytic activities of the proteasome such as the ATPase activity and substrate proteolysis are either stimulated or suppressed in the presence of DUBs. Cryo-EM structure of the proteasome in the act of substrate deubiquitylation and crystal structures of ubiquitin-DUB complexes, along with other cryo-EM studies of DUB in either proteasome subcomplexes or in the absence of substrates, have provided mechanistic insights into DUB activation and regulation by the substrate-proteasome interactions.
Intrinsic Deubiquitinase RPN11
RPN11, a JAMM metalloprotease, is the only intrinsic DUB stably associated with the proteasome and guarding the entrance of the central substrate-translocation channel in the base (Verma et al. 2002). RPN11 is an essential subunit of a functional proteasome and is crucial for cell viability (Maytal-Kivity et al. 2002; Guterman and Glickman 2004). Its role in the proteasome closely resembles the NEDD8 isopeptidase CSN5 in the COP9 signalosome (Cope et al. 2002). RPN11 resides immediately above the OB-ring of the AAA-ATPase motor and is intimately surrounded by the ubiquitin receptors RPN1 and RPN10. Its ubiquitin-binding site and the auxiliary ubiquitin-binding sites in the RPT4-RPT5 CC domain constitutes a concaved surface resembling a “hook”, which is structurally poised for capturing polyubiquitin chains (Dong et al. 2019; Chen et al. 2020). Proximity of RPN11 to the OB-ring and its geometric relationship to the surrounding ubiquitin receptor sites sterically preclude cleavage between ubiquitin moieties within polyubiquitin chains (Dong et al. 2019). Instead, it cleaves ubiquitin chains en bloc by hydrolyzing the isopeptide bond linking the C terminus of the first ubiquitin moiety to the substrate lysine (Yao and Cohen 2002).
The MPN domain of DUB RPN11 features three key regulatory regions: Insertion 1 (Ins-1), Insertion 2 (Ins-2) and a catalytic loop. The catalytic Zn2+ ion is coordinated with a metal-binding motif (EXnHXHX10D) and is surrounded by the Ins-1 region and the catalytic loop (Verma et al. 2002; Yao and Cohen 2002). The mutations His109Ala and His111Ala at the catalytic site do not impair the proteasome assembly, but inhibit substrate degradation and are cytotoxic in yeast (Verma et al. 2002). The Ins-1 region is located in the vicinity of the RPT4-RPT5 CC domain but does not make direct contact in the resting state of the proteasome. Ins-2 is notably flexible and is not well resolved in the crystal structures of the RPN11-RPN8 MPN-domain dimer (Pathare et al. 2014; Worden et al. 2014). By contrast, Ins-2 assumes an ordered structure and interacts with the toroidal domain of RPN2 in the proteasome (Dong et al. 2019). The catalytic loop of RPN11 contacts RPT4, forming hydrophobic interactions with an OB domain pore loop. These two inter-subunit interactions are critical to stabilize the conformations of Ins-2 and the catalytic loop in the pre-engaged state of the proteasome.
The lid subunit RPN8 and RPN11 form a heterodimer in the proteasome and when purified in isolation. In the free RPN11-RPN8 dimer, the Ins-1 region of RPN11 adopts a loop conformation that blocks the catalytic active site and self-inhibits the DUB activity (Worden et al. 2014; Pathare et al. 2014). In the free lid subcomplex, RPN5 further stabilizes the Ins-1 loop in the inhibited state, preventing RPN11 activation until it is assembled into the proteasome (Dambacher et al. 2016). In contrast, several related DUBs, such as Sst2 and AMSH-LP, remain active in isolation and their Ins-1 regions adopt a β-hairpin conformation, making the active site accessible (Sato et al. 2008; Davies et al. 2011; Shrestha et al. 2014). RPN11 retains a basal level of DUB activity in the context of the free RP subassembly (Lu et al. 2017b). In the free RP, RPN11 blocks the substrate entrance of the OB ring similar to its conformation in state ED of the proteasome, which prevents the OB ring from accepting substrates. The free RP exhibits a moderate ATP-independent DUB activity and its lack of any substrate-unfolding activity (Lu et al. 2017b). The ‘closed’ OB-ring in the free RP minimizes this risk of premature substrate deubiquitylation and unfolding prior to incorporation of the free RP into the mature proteasome. It remains to be clarified what cellular roles the basal DUB activity of RPN11 will play in the free RP.
Mechanism of Substrate Deubiquitylation by RPN11
The atomic structure of the substrate-bound proteasome in state EB reveals how the DUB activity of RPN11 is dependent on ATP hydrolysis and is coupled to substrate unfolding (Fig. 1.18) (Dong et al. 2019). In state EB, the ubiquitin-bound RPN11 interacts with both RPN8 and RPT5, which forms a subcomplex. This quaternary interface already starts to form in state EA2, when the substrate is not yet captured by the ATPase ring. Comparison of ubiquitin densities between states EA1, EA2 and EB suggests that the ubiquitin directly linked to the substrate is transferred from RPT4-RPT5 CC to RPN11 (Dong et al. 2019). In both states EA2 and EB, RPN11 does not interact with the substrate on the peptide-proximal side of the scissile isopeptide bond, explaining its non-specificity of removing ubiquitin chains from any ubiquitylated substrates.
Mechanism of substrate deubiquitylation by the 26S proteasome (PDB ID: 6MSE) (Dong et al. 2019). a Cryo-EM density maps of substrate-bound human proteasome in state EB at 3.3 Å. b A close-up view of the quaternary interface around the scissile isopeptide bond between ubiquitin and the substrate lysine in EB. The cryo-EM density is rendered as a transparent surface, superimposed with the cartoon representation of the atomic model. c A close-up view of the zinc ion (hotpink sphere) closely approached by the isopeptide bond. The zinc ion density is shown as a blue mesh at 10σ level. Side chains of Rpn11 coordinating with the zinc ion are labeled
The ubiquitin-binding interface is located at a hydrophobic pocket around Trp111 and Phe133 of RPN11 (Worden et al. 2014; Pathare et al. 2014; Dong et al. 2019). The Ins-1 loop adopts a β-hairpin conformation, and pairs from one side with the C-terminal strand of ubiquitin and from the other side with a segment of the RPT5 N-loop (residues 99–119) emanating from the OB ring. Together they form a four-stranded β-sheet. This β-sheet places the isopeptide bond right next to the zinc ion in the RPN11 active site (Dong et al. 2019). The Ins-1 β-hairpin is rotated outward in state EB relative to its conformation in the crystal structure of ubiquitin-bound RPN11 and is better aligned with the zinc-binding site in the proteasome (Worden et al. 2017). The conserved RPT5 N-loop stabilizes the ubiquitin contact with RPN11 and adjusts the orientation of the isopeptide bond for efficient deubiquitylation, whereas it is unstructured in most other states (EA1, EC1, EC2, ED1 and ED2).
The Ins-1 region of RPN11 keeps changing its conformation with different states of the proteasome (Fig. 1.19). The Ins-1 remains as a β-hairpin conformation throughout states EA2, EB and EC1, whenever ubiquitin is bound. It becomes a large open loop in state EA1 and refolds into a closed, tighter loop in states EC2, ED1, and ED2 whenever ubiquitin is released. The quaternary organization surrounding the zinc-binding site of RPN11 appear to explain why the DUB activity of RPN11 is prominently upregulated in the proteasome than in its non-proteasome forms, such as the free RP assembly (Lu et al. 2017b), the free lid subcomplex (Dambacher et al. 2016) and the heterodimeric RPN8-RPN11 form (Pathare et al. 2014; Worden et al. 2014). The dynamic regulation of Ins-1 conformations allows allosteric coordination and timing between substrate commitment, deubiquitylation and degradation. Initial substrate engagement with the AAA-ATPase motor allosterically regulates the conformational changes of RPN11 and RPT5 N-loop and enables deubiquitylation only for committed substrates. Indeed, disrupting the closed state of Ins-1 stimulates the RPN11 DUB activity but also causes degradation failure, because ubiquitin chains are prematurely removed, allowing substrates to escape from the proteasome prior to translocation and degradation (Worden et al. 2017).
Conformational dynamics of the RPN11 associated with an enhanced isopeptidase activity in the proteasome (PDB IDs: 6MSB, 6MSD, 6MSE, 6MSG, 6MSH, 6MSJ, 6MSK) (Dong et al. 2019). a Overview showing the inter-subunit relationship between RPN2-RPN11-RPN8 and the CC-OB domains of the ATPases in state SA. Black box indicates the location to which the close-up view in panel (b) is zoomed. b Close-up view of Rpn11 in cartoon representation. The adjacent RPT subunits and RPN2 are shown in surface representation. c Close-up view of the Zn2+ active site of Rpn11. The viewing angle is rotated clockwise ~90° relative to that of panel (E). d Comparison of the Ins-1 loop of RPN11 in different states. e Comparison of the RPN11 structures in states EA2, EB and EC1 around the zinc-binding site and Ins1 region with that in the crystal structure (PDB ID: 5U4P) of ubiquitin-bound RPN11-RPN8 complex from yeast (Worden et al. 2017). f Closeup comparison of the RPN11 Ins1 structure between state EB and 5U4P (left two panels) and between state EC1 and 5U4P in two orthogonal perspectives, showing a 5-Å displacement of the Ins-1 β-hairpin in EB relative to 5U4P or EC1. This displacement is not observed between EC1 and 5U4P, suggesting that the Ins-1 β-hairpin tilt in EB is mostly to optimize the coordination of isopeptide bond with the zinc ion
The structural revelation of state EB fills up a fundamentally critical intermediate, allowing the observation of stepwise activation of the CP by docking of the RPT C-termini into the α-pockets one at a time (Dong et al. 2019). During the process of CP gate opening, the relative positions of the RPN11 catalytic site, the central pore of the ATPase ring, and the CP gate are coaxially aligned (Matyskiela et al. 2013; Sledz et al. 2013; de la Pena et al. 2018; Dong et al. 2019), preparing the proteasome for processive substrate translocation into the CP chamber.
Deubiquitinase USP14
USP14 or its yeast ortholog Ubp6 is a cysteine-dependent DUB that is reversibly associated with the proteasome (Borodovsky et al. 2001). The N-terminal UBL domain of USP14 binds RPN1, whereas its catalytic USP domain contacts the exterior of the OB and AAA rings opposite RPN11 (Bashore et al. 2015; Shi et al. 2016; Huang et al. 2016; Aufderheide et al. 2015; Leggett et al. 2002). Such multivalent interactions facilitate stimulation of its DUB activity by lifting off the inhibitory loops blocking the active site. Thus, the DUB activity of USP14 is allosterically regulated by its interaction with the proteasome and is tightly coupled with the conformational state of the proteasome (Bashore et al. 2015; Hanna et al. 2006). Association with the proteasome activates USP14/Ubp6 ∼300- to 800-fold when assayed with the model substrate ubiquitin-AMC (Leggett et al. 2002; Lee et al. 2010). Re-equilibrating the yeast proteasome conformation toward the open-gate states with ATPγS further increases the activity of Ubp6 by a couple of folds in cleaving the ubiquitin-AMC substrate (Bashore et al. 2015). The active sites of USP14 and RPN11 are only 35 Å apart in the base, allowing USP14 to antagonize ubiquitin binding to RPN11 (Bashore et al. 2015; Hanna et al. 2006). Ubiquitin-bound USP14 appears to stimulate the proteasomal ATPase activity and CP gate opening, suppress substrate degradation by destabilizing the resting state of the proteasome, inhibit deubiquitylation by RPN11 and the basal peptide hydrolysis, and surprisingly enhance the E4 ligase UBE3C/Hul5 association with the proteasome (Bashore et al. 2015; Aufderheide et al. 2015; Peth et al. 2009; Leggett et al. 2002; Kuo and Goldberg 2017; Kim and Goldberg 2017). Curiously, the UBL domain of USP14 alone can stimulate the proteasome activation (Kim and Goldberg 2018). Hence, USP14/Ubp6 may allosterically regulate the proteasome activities at multiple levels, including the recognition of ubiquitin or substrate, timing of deubiquitylation and processivity of substrate proteolysis.
When the USP domain is recruited to the base of the RP, the OB-ring is adjacent to the proximal ubiquitin binding site of USP14 (Huang et al. 2016). This resembles the quaternary structural relationship between the OB-ring and, in its immediate vicinity, the proximal ubiquitin-binding site on RPN11 (Dong et al. 2019). Thus, the biochemical behaviors of RPN11 and USP14 with respect to an incoming substrate might be comparable and somewhat competitive to each other. With a preference for cleavage at the substrate-linked isopeptide of ubiquitin chains, they both could reverse ubiquitin modifications en bloc on a time scale of milliseconds to seconds (Lee et al. 2016). Interestingly, USP14 can rapidly cleave ubiquitin from a substrate that is conjugated to multiple polyubiquitin chains, whereas deubiquitylation of single-chain conjugates or disassembly of free polyubiquitin chains by USP14 is very slow (Lee et al. 2016). Curiously, USP14 can cleave long, unbound Lys48-linked chains better than does RPN11 for certain substrates (Mansour et al. 2015). Thus, USP14 seems to be specialized in removing supernumerary ubiquitin chains and cannot replace RPN11 in thorough deubiquitylation of substrates (Lee et al. 2016). In contrast to the essential role of RPN11 in proteasomal activities, inhibition or absence of USP14/Ubp6 in the proteasome accelerates substrate degradation in vitro (Hanna et al. 2006) and is tolerable for yeast cells (Guterman and Glickman 2004). However, the absence of Ubp6 causes a growth deficiency due to enhanced proteasomal degradation and depletion of free ubiquitin (Leggett et al. 2002). USP14 deficiency in mouse embryonic fibroblasts increased the abundance of UCH37 and RPN13 in the proteasome, suggesting that UCH37 may compensate the loss of USP14 (Kim and Goldberg 2017).
Deubiquitinase UCH37
UCH37 is another cysteine-dependent DUB reversibly associated with the proteasome in higher eukaryotes, such as Homo sapiens and Drosophila melanogaster. Its ortholog Uch2 was found in Schizosaccharomyces pombe but not Saccharomyces cerevisiae (Lam et al. 1997; Holzl et al. 2000). UCH37 consists of two domains, an N-terminal catalytic domain representative of the UCH family and a fibrous C-terminal tail (Burgie et al. 2012). UCH37 is activated by binding of its C-terminal tail to the N-terminal DEUBAD domain of RPN13 in the proteasome but not in a free UCH37/RPN13 complex (Yao et al. 2006; Sahtoe et al. 2015; Vander Linden et al. 2015). A loop sequesters the catalytic cysteine (Cys88) in the free UCH37 (Burgie et al. 2012). Upon association with RPN13, this loop is remodeled to open the access to the catalytic triad residues (Cys88, His164 and Asp179), which gives rise to an approximately five-fold increase in its affinity for ubiquitin (Yao et al. 2006; Hamazaki et al. 2006; Qiu et al. 2006; Sahtoe et al. 2015; Vander Linden et al. 2015). Within the free RPN13, the DEUBAD domain interacts with the PRU domain and sterically occludes its ubiquitin-binding site. Association of RPN13 with RPN2 in the proteasome modifies this inter-domain interaction and makes the PRU domain to become accessible for ubiquitin binding, leading to activation of UCH37 (Chen et al. 2010; Lu et al. 2017a).
Once activated in the proteasome, UCH37 can catalyze the removal of distal Lys48-, Lys11-, and Lys6-linked ubiquitin chains conjugated to substrates (Lam et al. 1997). Inadequately ubiquitylated proteins could be released by UCH37 from the proteasome. UCH37 may also reverse regulatory ubiquitylation modifications from certain proteasome subunits (Jacobson et al. 2014). It might trim distal ubiquitin moieties, release them from the ubiquitin receptors and vacate the ubiquitin-binding sites in the proteasome (Zhang et al. 2011). Despite its putative location that is about the length of tetraubiquitin away from the OB ring of the AAA-ATPase motor (Holzl et al. 2000; Dong et al. 2019; Lu et al. 2020), UCH37 association with the proteasome can allosterically stimulate the proteasomal ATPase activity and CP gate opening (Peth et al. 2013a). These allosteric effects are in line with those seen in the USP14-bound proteasome and further underscore the global allosteric coupling between all components of the proteasome. In addition to its role in the proteasome, UCH37 also appears as a subunit of the chromatin-remodeling complex INO80 (Yao et al. 2008). It remains to be clarified what substrate specificities of UCH37 are and how UCH37 acts on its substrates in the proteasome. How the three DUBs, RPN11, USP14 and UCH37, are coordinated and collectively regulated in the context of the proteasome will be an intriguing open question awaiting answers.
Operating Principles of Proteasomal AAA-ATPase Motor
Proteasomal AAA-ATPase Motor
The proteasome utilizes its ATPase motor module to mechanically unfold the globular domains of substrates and deliver them into the CP for their breakdown into short polypeptides. The proteasomal ATPases are members of the classic AAA clade in the AAA+ superfamily (Fig. 1.20) (Vale 2000; Smith et al. 2006). Other members in this clade include FtsH family, Cdc48 family, and ClpA/B/C-Domain 1 (D1) family (Ogura and Wilkinson 2001; Lupas and Martin 2002; Iyer et al. 2004; Erzberger and Berger 2006; Hanson and Whiteheart 2005). The HCLR clade, which includes the HslU/ClpX family, ClpA/B/C-Domain 2 (D2) family, and Lon family, and features a pre-sensor 1 insertion residing in at least one AAA+ module, is found in many protease complexes and functions as substrate unfolding machinery resembling the proteasomal AAA-ATPase motor (Sauer and Baker 2011).
Asymmetric structure of the ATPase heterohexamer in the SA/EA state (PDB ID: 6MSB) (Chen et al. 2016a; Dong et al. 2019). a The OB-AAA domain structures of RPT subunits in a cartoon representation from a perspective showing an L-shape. The leftmost panel shows the six RPT structures superimposed, with each structure separately shown on the right. b The dimeric AAA domain structure of two adjacent RPT subunits. The leftmost panel shows the six AAA dimer structures superimposed together, with each structure separately shown on the right. c The electrostatic structure of the OB ring (left), AAA ring (middle), and ATPase channel (right). d The inter-subunit interfacial areas between two adjacent RPT subunits. The black numbers show the total inter-subunit interfacial areas, whereas the red numbers show the interfacial areas between the adjacent AAA domains. e A table shows the inter-subunit interfacial areas between the large and small AAA subdomains from two adjacent RPT subunits. Note that the two adjacent small AAA subdomains do not contact each other
The AAA domain of RPT contains highly conserved motifs commonly observed in the AAA+ superfamily proteins, including Walker A, Walker B, inter-subunit signaling (ISS) motif, sensor 1, arginine finger (R-finger), sensor 2, and pore-1/2 loops (Fig. 1.21) (Sauer and Baker 2011; Ogura and Wilkinson 2001; Lupas and Martin 2002; Iyer et al. 2004; Hanson and Whiteheart 2005; Erzberger and Berger 2006; Chang et al. 2017; Zhang and Mao 2020; Puchades et al. 2020). The nucleotide-binding pocket is intimately surrounded in “cis” by Walker A, Walker B, sensor 1, and sensor 2 within the same AAA domain, and two R-fingers and ISS motif in “trans” from the large AAA subdomain of the clockwise adjacent ATPase subunit (Wendler et al. 2012). With these structural motifs, ATP hydrolysis is coupled to conformational changes of the ATPases that apply mechanical work on a substrate through the axial channel-aligning pore loops.
Conformational Dynamics of the AAA-ATPase Motor
The Substrate Translocation Pathway
The substrate-translocation pathway in the proteasomal ATPase motor is narrowed by the inward-facing pore loops that are common to the classic clade ATPases (Fig. 1.22) (Erales et al. 2012; Beckwith et al. 2013; Hinnerwisch et al. 2005; Martin et al. 2008; Dong et al. 2019). The AAA channel exhibits a right-handed helical architecture with a much narrower constriction compared to the OB channel in the resting state (SA) of the proteasome. The interior of the AAA channel is largely negatively charged, in contrast to the positively charged interior of OB channel (Chen et al. 2016a; Dong et al. 2019). This may reflect distinct roles of the OB and AAA domains in priming and mechanically translocating substrates. Despite this difference, the OB and AAA channels share a dramatic enrichment of tyrosine residues. The OB channel features six tyrosine residues (Tyr147 in RPT1, Tyr72 and Tyr121 in RPT6, Tyr111 in RPT3, Tyr79 in RPT4, and Tyr158 in RPT5), whose sidechain oxygen atoms point towards the substrate-translocation pathway. Likewise, the AAA channel is also decorated with five tyrosine residues in the resting state. These tyrosine residues may facilitate substrate engagement and translocation via cation-π or π-π interactions (Glynn et al. 2009).
Architecture of the substrate-translocation channel in the human proteasome at a resting state (PDB ID: 6MSB) (Chen et al. 2016a; Dong et al. 2019). a Top view of the pore loops aligning along the channel axis from the perspective of the OB domain. The solvent-accessible surface of the channel calculated by the HOLE program is rendered by surface dots. b Side views of the pore-1 loops from six RPT subunits decorating the channel, which align along the channel in a spiral staircase formed from RPT1-RPT5, with a backward recession in RPT6 pore-1 loop that is slightly away from the major channel pathway. The pore-1 loops form a helical part of the channel interior as illustrated by the dashed green line. c Side view of the pore-2 loops from six RPT subunits decorating the channel, which form another spiral staircase. These pore-2 loops form another helical part of the channel interior, illustrated by the red dashed line. d Side view of the complete ATPase channel including components from both the OB and AAA domains, calculated by the HOLE program (Smart et al. 1996). Side-chain patterns observed along the substrate-translocation pathway are highlighted. The five tyrosine residues, highlighted by transparent sphere representation, and a number of hydrophobic and negatively charged residues decorate the AAA channel; color codes for ATPase protomers match those shown in (a). Lower left inset, a schematic cartoon showing that the pore-2 loops of RPT3, RPT4, RPT5 pair laterally with the pore-1 loops of RPT4, RPT5 and RPT1, respectively, to form the three narrowest constrictions in the AAA channel in the resting state of the proteasome
The pore-1 loops of the RPT subunits feature the conserved ‘[Tyr/Phe]-[Val/Leu/Ile]-Gly’ sequence pattern and arrange into a super-helical staircase. In many ATP-dependent AAA unfoldases, such as ClpX, HslU, LonA, FtsH and PAN (Glynn et al. 2009; Iosefson et al. 2015), their homologous pore-1 loops drive substrate translocation. The pore-2 loops constitute a second super-helical staircase running in the opposite of the pore-1 loop staircase (Fig. 1.22), which may function a ‘sensor’ for the state of the substrate translocation as these pore-2 loops are directly extended from the Walker B motif and can translate the presence of substrate in the channel into the activation of ATP hydrolysis, by modulating the distance between the γ-phosphate of ATP and the Walker B motif (Zhang and Wigley 2008). In contrast to the hydrophobicity of the pore-1 loops, the pore-2 loops are heavily decorated with negatively charged residues, including seven glutamates and two aspartates. The markedly contrasting side-chain patterns in the two complementary parts of the channel surfaces indicate that they play somewhat different roles in substrate translocation.
The AAA channel exhibits three constrictions in the resting state of the proteasome, featuring the positional overlap along the channel axis between the two pore-loop staircases. The aromatic and hydrophobic residues in the pore-1 loops from RPT4, RPT5, and RPT1 are equilaterally paired with adjacent charged residues in the pore-2 loops of RPT3, RPT4 and RPT5, respectively (Fig. 1.22d). Interestingly, mutations in the pore loops resulted in similar phenotypic effects between each pore loop pair. For pore-1 loop, RPT4 has the strongest phenotype, whereas for pore-2 loop the partner RPT3 has the strongest phenotype (Beckwith et al. 2013). The strength of phenotype is also similarly reduced with their distance along the pore axis. In the resting state, the channel radius is constricted to as little as ~2 Å, which must be opened up to allow the insertion of a substrate (Dong et al. 2019). The pore loops of RPT6 is uniquely displaced from the channel axis and does not contribute to these constrictions. Consistently, the pore-2 loop mutations of RPT6 were shown to have little impact on substrate degradation rates (Beckwith et al. 2013).
Dynamic Substrate Interactions with the AAA Ring
The AAA channel formed by the pore loops is structurally plastic (Eisele et al. 2018; Zhu et al. 2018). Even without a substrate, the pore-loop architecture in the states with an open CP are dramatically reorganized between super-helical staircase and saddle-shaped circles deviating from a rigorous staircase arrangement, as typically characterized in state SD1, SD2 and SD3 (Zhu et al. 2018). While the substrate-free conformation of the ATPase ring in state SD2 resembles that of the substrate-bound one in state ED2, the pore-loop conformation is relaxed and less organized in the absence of the substrate (Zhu et al. 2018; Dong et al. 2019). The plasticity of the pore-loop conformation allows the AAA channel to not only accommodate a vast diversity of substrate sequences, but also a hairpin-like polypeptide loop as typically observed in the cryo-EM structure of AAA+ ATPase Vps4 bound with a circular substrate (Han et al. 2019).
In state EB, the substrate density is approximately centered inside the OB ring (Dong et al. 2019). By contrast, the substrate closely approaches Phe118 of RPT1 inside the OB ring in states EC1, EC2, ED1 and ED2. Within the AAA channel, the substrate is gripped by the aromatic residues of pore-1 loops arranging into a right-handed spiral staircase (Fig. 1.23). Either tyrosine or phenylalanine in the pore-1 loops intercalates with the fully stretched main chain of the substrate through hydrophobic interactions. The main chains of the pore-1 loops potentially form hydrogen bonds with the substrate main chain. The adjacent pore-1 loops are spaced by two amino acid residues of the substrate, thus evenly distributed along the substrate (de la Pena et al. 2018; Dong et al. 2019). This “two-residue spacing” appears to be a highly conserved structural “rule” for substrate-pore loop interactions among many other AAA+ ATPases, such as p97/Cdc48 (Ripstein et al. 2017; Olszewski et al. 2019), FtsH-like AAA proteases (Puchades et al. 2017, 2019) and Hsp104 disaggregase (Gates et al. 2017). This perhaps implies a common mechanism for the force generation by nonspecific, intercalated stacking interactions between the pore loop’s aromatic side chains and the stem of substrate side chains.
Dynamic substrate-pore loop interactions (PDB IDs: 6MSE, 6MSG, 6MSH, 6MSJ, 6MSK) (Dong et al. 2019). a Varying architecture of pore-1 loop staircase interacting with the substrate in five states (EB, EC1,2 and ED1,2). Aromatic residues in the pore-1 loops are labeled and shown in stick representation superimposed with transparent sphere representation for highlighting. The distances from disengaged pore-1 loops to the substrate are marked. b Side view of all substrate-bound ATPase-RPN11 structures superimposed together based on structural alignment against the CP. c A diagram summarizing the axial stepping of the substrate-contacting pore-1 loops and their correlations with nucleotide states. The vertical axis shows the relative location of pore-1 loops interacting the substrate, with the CP positioned at the bottom. The relative distance from the lowest substrate-pore loop contact is labeled using the number of residues as a metric. State EC1 is omitted here as its AAA-ATPase structure is identical to that of EC2. The color code of subunits used in all panels is shown in the upper right inset
The architectural organizations of the substrate-pore loop staircase from states EB to ED2 are all quite comparable, despite different subunit combination. The highest positions in contact with the substrate in states EB, EC, ED1 and ED2 are occupied by the pore-1 loops of RPT3, RPT6, RPT1 and RPT5, respectively. Notably, the pore-1 loop of RPT3 is translated from one end to the other end in the substrate-pore loop staircase during state transitions from EB to ED2. Meanwhile, the pore-2 loops form another shorter staircase and support the substrate from the opposite of the pore-1 loop staircase (Chen et al. 2016a; Zhu et al. 2018; de la Pena et al. 2018; Dong et al. 2019). In states EB, ED1 and ED2, the pore loops of only one ATPase subunit (RPT6, RPT5 and RPT4, respectively) are dissociated from the substrate, whereas the pore loops of two ATPase subunits (RPT1 and RPT2) are displaced from the substrate in states EC1 and EC2. Thus, asymmetric substrate interactions with the pore loops would inevitably exert an allosteric impact on the overall asymmetric ATPase motor conformation. Consistently, functional asymmetry of the six ATPases in substrate degradation has been observed in mutagenesis experiments (Beckwith et al. 2013; Tian et al. 2011; Erales et al. 2012).
Nucleotide-Binding Sites
Three distinct nucleotide states in the proteasomal ATPase subunits have been observed, i.e., ATP-bound, ADP-bound, and apo-like states. The apo-like state is referred to the absence or a very weak density of nucleotide in the nucleotide-binding pocket (Dong et al. 2019). The nucleotide-binding site is located at the Walker A motif located near a short loop between the small and large AAA subdomains (Fig. 1.24). The nucleotide state controls the ATPase conformation by modifying the inter-domain and inter-subunit interactions (Sauer and Baker 2011; Ogura and Wilkinson 2001; Sledz et al. 2013). The proteasome hydrolyzes ATP slowly in the absence of substrate, on average only 27 ATPs per minute per complex (Hoffman and Rechsteiner 1996; Kim et al. 2015; Smith et al. 2011; Benaroudj et al. 2003). ATP hydrolysis in RPT subunits requires magnesium ion bound to the β- and γ-phosphates. The pre-engaged proteasome, despite being saturated with ATP, stays in a basal resting state of minimal ATPase activity (Benaroudj et al. 2003; Peth et al. 2013b; Smith et al. 2011; Kim et al. 2015). Substrate engagement stimulates ATP hydrolysis by tightening the overall ATPase ring structures, by initiating highly concerted intersubunit interactions and by coordinating conformational changes around the ring (Dong et al. 2019).
Nucleotide-binding sites in the substrate-bound human proteasome in distinct states (PDB IDs: 6MSB, 6MSD, 6MSE, 6MSG, 6MSH, 6MSJ, 6MSK) (Dong et al. 2019). Comparison of the nucleotide-binding pockets of six ATPases in all states illustrates a common pattern in the geometry of the nucleotide-binding sites. Each row shows the geometry of the nucleotide-binding pocket of one ATPase in all six states. In each panel showing an ATP or ADP-bound state, one red dashed line marks the distance from β/γ-phosphate of nucleotide to the arginine finger of the adjacent ATPase, while the other line marks the distance from the same phosphate to the Walker B motif. In the case of apo-like states, the red lines extend to the proline of the Walker A motif rather than to the phosphate groups. These geometries indicate the potential reactivity of these sites (Zhang and Wigley 2008). When the ATPase is positioned in the middle of pore-loop staircase, but not at the lowest position, the nucleotide-binding pockets are tightly packed whenever ATP or ADP is bound. By contrast, when the ATPase is either in the lowest position of the substrate-pore loop staircase or disengaged from the substrate, the nucleotide-binding pocket is open regardless of whether it is ADP-bound or free of nucleotide. The resolution of 2.8–3.6 Å in the cryo-EM maps of substrate-bound human proteasome allows unambiguous assignment of nucleotide densities to ADP and ATP in the nucleotide-binding pockets of the ATPases. Except for EA, at least one ATPase subunit in each conformational state exhibits a very weak or partial density for a potential nucleotide in its nucleotide-binding pocket, which precludes de novo atomic modeling of nucleotide coordinates into the density of that subunit. The nucleotide state of these ATPases has been referred to as an apo-like state. It is not possible to differentiate between ATP and ATPγS at the present resolution. Because the mixture of both in the particle population, the 3D classification is not supposed to differentiate ATP against ATPγS at the present resolution. For simplicity, the potential nonhydrolyzed nucleotide has been modelled with ATP
The nucleotide states of RPT subunits in the substrate-engaged proteasome exhibit common features reflecting a marked spatiotemporal continuity of the AAA-ATPase motor (Fig. 1.24). Comparison of the pre-engaged structure with the substrate-engaged ones suggests that ATP hydrolysis occurs first in RPT6, followed by hydrolytic events in RPT5 (Dong et al. 2019; Schweitzer et al. 2016; Huang et al. 2016; Chen et al. 2016a). Notably, the ADP-bound states navigate counterclockwise sequentially from RPT6 to RPT3 throughout all six ATPase subunits, indicating a full cycle of coordinated ATP hydrolysis around the AAA-ATPase ring from state EA to ED2. Magnesium ion density is evident next to ATP and, in fewer cases, also to ADP as well in the cryo-EM density maps of several states (Dong et al. 2019). Whenever the two adjacent ATPase subunits are both in contact with the substrate, their intersubunit interface is stabilized by the insertion of a phenylalanine residue from the ISS motif of the clockwise subunit into a phenylalanine-containing pocket of the counterclockwise subunit near its nucleotide-binding site and pore-1 loop (Chang et al. 2017; Puchades et al. 2017; Dong et al. 2019). This ISS-mediated interaction locks the substrate-bound ATPase subunits together into an approximate rigid body, a mechanical property that is likely required to sustain the translocating force applied by the pore loops on the substrate. It remains to be clarified how intersubunit communications by the ISS motif help to order the timing of ATP hydrolytic events around the ring during substrate translocation.
Substrate Interactions Coupled with ATP Hydrolysis
Key structural motifs of each ATPase subunit couple its interactions with the substrate with the ATP hydrolytic cycle (Ogura and Wilkinson 2001; Lupas and Martin 2002; Iyer et al. 2004; Hanson and Whiteheart 2005; Erzberger and Berger 2006; Sauer and Baker 2011). In other words, the nucleotide states determine the geometry of the nucleotide-binding pockets and are strongly coupled with the substrate-pore loop interactions (Dong et al. 2019). This coupling is reflected in several lines of structural consensus. First, the apo-like state is always observed in the substrate-disengaged ATPases. All apo-like subunits form prominent gaps at their inter-subunit interfaces with their nearest neighbors on both sides. Specifically, the ISS motifs of these apo-like subunits are in a retracted conformation and far away from its adjacent subunit. Likewise, the arginine fingers from the adjacent subunit fall more than 10 Å apart from the Walker A motif of the apo-like subunits, leaving an apparently open nucleotide-binding pocket. Second, the substrate-engaged ATPase that is closest to the CP is always found to be bound with ADP. The clockwise nearest neighbor of the ADP-bound subunit is always found to be in an apo-like state. Third, whenever the substrate-engaged ATPases resides in the middle or top registry in the substrate-pore loop staircase, the nucleotide-binding pocket is always closed by the arginine fingers of the adjacent ATPase. Except states EA and EB, these closed nucleotide-binding sites are always occupied by ATP.
The AAA domain remains most stable when bound to ATP. Under this circumstance, the entire AAA domain may still rotate as a rigid body. Systematic structural comparison has revealed that ADP release is associated with the largest hinge-like inter-domain rotation of 15–25° between its small and large AAA subdomains (Fig. 1.25) (Dong et al. 2019). Release of γ-phosphate after ATP hydrolysis does not immediately trigger inter-domain motion of the AAA domain, as the ATPase conformation is also regulated by the inter-subunit interactions. The subsequent disengagement of the ATPase from the substrate facilitates the ADP release, liberates the potential energy that is converted into kinetic energy driving the hinge-like inter-domain rotation. Dissociation from the substrate also allows the ATPase to flip outward by 30–40° relative to the entire ATPase ring. The kinetic energy is liberated and spread out to drive rigid-body rotation of four or five substrate-bound ATPase subunits, propelling the substrate toward the CP (de la Pena et al. 2018; Dong et al. 2019). These structural findings are in line with a sequential model of ATP hydrolysis around the ATPase ring (Smith et al. 2011; Kim et al. 2015; Olivares et al. 2018). Notably, a common feature observed in most other substrate-bound ATPase hexamer structures is that at least one subunit is dissociated from the substrate for nucleotide exchange (Gates et al. 2017; Puchades et al. 2017; Monroe et al. 2017; Ripstein et al. 2017; Deville et al. 2017; Alfieri et al. 2018; Thomsen and Berger 2009; Han et al. 2017; Puchades et al. 2019).
Asymmetric intramolecular dynamics of AAA domains driven by nucleotide exchange (PDB IDs: 6MSB, 6MSD, 6MSE, 6MSG, 6MSH, 6MSJ, 6MSK) (Dong et al. 2019). Superposition of the AAA domain conformations of the same subunit RPT6 (a), RPT1 (b), RPT2 (c), RPT3 (d), RPT4 (e) or RPT5 (f) from distinct states aligned against their large AAA subdomains. Superposition of the RPT6 AAA domain structures from five distinct states aligned against the large AAA subdomain shows that RPT6 assumes three major conformations. As shown in the boxed inset, transition from EA to EB involves both refolding of the pore-2 loop, shown in the right insert, and a 20° rigid-body rotation between the large and small AAA subdomains. RPT1, RPT4, and RPT5 assume two major conformations between apo-like and nucleotide-bound states, while RPT2 assumes three major conformations
Principles of Coordinated ATP Hydrolysis in Functional Regulation
The proteasomal ATPase motor bears a greater degree of inter-subcomplex interactions than most of other AAA+ ATPase motors. On one side, the ATPase motor interfaces with many RPN subunits in the lid and base. On the other side, it forms a highly dynamic, multivalent interface with the α-ring. Such a structural complexity is presumably evolved to achieve the functional complexity of the proteasome. Unexpectedly, three distinct modes of coordinated ATP hydrolysis in the proteasomal ATPase ring appear to regulate the intermediate functional steps of the proteasome (Fig. 1.26) (Dong et al. 2019). The capability of functioning in multiple modes implies the existence of multiple feasible pathways of conformational transitions in the same AAA-ATPase hexamer induced by coordinated ATP hydrolysis.
Principal modes of coordinated ATP hydrolysis in the functional regulation of the proteasome (Dong et al. 2019). The cartoon summarizes the concept of three principal modes of coordinated ATP hydrolysis observed in the seven states and the proposed model of how they regulate the complete cycle of substrate processing by the proteasome holoenzyme. Coordinated ATP hydrolysis in Modes 1, 2 and 3 features hydrolytic events in two oppositely positioned ATPases, in two consecutive ATPases, and in only one ATPase at a time, respectively. Substrate processing undergoes three major steps before CP gate opening for processive translocation: (1) ubiquitin recognition; (2) simultaneous deubiquitylation and substrate engagement with the AAA-ATPase ring; and (3) translocation initiation, which involves multiple simultaneous events, including ubiquitin release, ATPase repositioning and switching of RPT C-tail insertion pattern. In some cases, the initiation of translocation may precede deubiquitylation. In steps 1 and 2, the ATPases follow the Mode-1 ATP hydrolysis. In step 3, they follow the Mode-2 ATP hydrolysis. After the gate is open, the AAA-ATPases hydrolyze ATP in Mode 3, in which only one nucleotide is hydrolyzed at a time
Mode 1 Regulates Ubiquitin Recognition, Initial Substrate Engagement and Deubiquitylation
Mode 1 is observed in states EA1, EA2 and EB and features coordinated ATP hydrolysis in a pair of oppositely positioned ATPases, which is associated with the steps of initial ubiquitin recognition and deubiquitylation (Peth et al. 2010; Worden et al. 2017; Dong et al. 2019). Before the proteasome gets ready to cleave the ubiquitin chain conjugated to the substrate, the ADP in RPT6 is released and the ATP in both RPT2 and its opposite subunit RPT4 are hydrolyzed. These events trigger refolding of a segment spanning residues 251–266 in RPT6 and drive a marked outward rotation of the entire RPT6 AAA domain (Fig. 1.25). Regulated by this key conformational change, the coordinated ATP hydrolysis in RPT5, and then in RPT4, is expected to relax the conformational rigidity of the AAA ring to enable an iris-like movement opening the AAA channel for substrate insertion (Fig. 1.27). The position of RPT6 at the seam of the AAA ring endows it with certain energetic advantage in allosterically triggering the AAA channel opening with little perturbation to the pore loop staircase structure, which is poised to accept an unfolded segment of the substrate without changing the overall ATPase architecture. This mode of coordinated ATP hydrolysis is reminiscent of the nucleotide states in the crystal structure of the ClpX protease, which drive different structural changes in the ATPase ring likely because of the absence of a substrate (Glynn et al. 2009).
Mechanism of initial substrate engagement with the proteasome (PDB IDs: 6MSB, 6MSD, 6MSE) (Dong et al. 2019). a Superposition of the AAA-ring structures of states EA (grey) and EB (color). The insets show side-by-side comparison of RPT6 conformations in the four most distant states. Interfacial gaps are marked by red dashed lines. b Pore-1 loop staircase before and after engaging with the substrate in state EB as compared to that in state EA1
Mode 2 Regulates CP Gating, Ubiquitin Release and Initiation of Substrate Translocation
Mode 2 is observed in states EC1 and EC2 and is characteristic of coordinated ATP hydrolysis in at least two adjacent substrate-disengaged ATPases, which is linked to the intermediate steps of CP gating, ubiquitin release and initiation of substrate unfolding (Dong et al. 2019). Upon deubiquitylation, the proteasome must prepare for substrate translocation and allosteric regulation of the CP gate opening. To this end, ATP hydrolysis in two adjacent subunits, RPT1 and RPT5, is coupled with disengagement of RPT2 and RPT1 from the substrate. As RPT6 acquires ATP again and re-binds the substrate at the top of the substrate-pore loop staircase, forward translocation of the substrate over a two-residue distance is driven by the conformational changes of ATPases from state EB to EC. During the following EC2-to-ED1 transition, RPT1 and RPT2 both need to re-bind ATP and the substrate and return to the top of the substrate-pore loop staircase, whereas RPT5 is about to release its ADP. As multiple events occur during this process, it is anticipated that several key intermediates were missed in cryo-EM analysis and it remains to be addressed how the EC2-to-ED1 transition is accomplished. Recent cryo-EM structures of the yeast Cdc48 and of FtsH-like mitochondrial protease AFG3L2 exhibited a substrate-bound ATPase architecture highly comparable to Mode 2 of the 26S proteasome (Twomey et al. 2019; Puchades et al. 2019).
Mode 3 Regulates Processive Substrate Unfolding, Translocation and Degradation
Mode 3 is observed in states ED1 and ED2 and features ATP hydrolysis in only one ATPase at a time during processive substrate unfolding and translocation (Fig. 1.28) (Dong et al. 2019). When the pore-1 loop of the substrate-engaged ATPase reaches the CP-proximal bottom of the substrate-pore loop staircase, the nucleotide state of the ATPase subunit is always ADP-bound. During the process of ADP release, the inter-domain rotation within the AAA domain flips the AAA domain outwards away from the rest of the ATPase ring and detaches its pore loops from the substrate. Meanwhile, its counterclockwise adjacent substrate-engaged ATPase is pushed to the bottom of the ATPase ring, whereas its clockwise neighboring ATPase, which was an apo-like detached seam earlier, now re-binds ATP and the substrate at the top of the ATPase staircase. In concert with these motions, the other three substrate-engaged, ATP-bound ATPases rotate downwards approximately as a rigid body driven by the intrinsic conformational changes in the substrate-disengaged ATPase undergoing nucleotide exchange. In support of coordinated ATP hydrolysis of Mode 3 in both human and yeast proteasome (de la Pena et al. 2018; Dong et al. 2019), five distinct conformations of the PAN ATPase ring in the archaeal PAN-CP complex in the absence of a substrate all exhibits only one ATPase disengaged from the rest of the ATPase ring, a key feature consistent with Mode 3 (Majumder et al. 2019). Both the Mode-2 and Mode-3 hydrolysis models confer a unidirectional propagation of conformational changes in the ATPase ring.
Mechanism of processive substrate translocation in the proteasome (PDB IDs: 6MSH, 6MSJ, 6MSK) (Dong et al. 2019). a Structural comparison between ED1 (color) and EC2 (grey), by using the large AAA subdomain of RPT5 to align the AAA ring structures in distinct states. A 30° out-of-plane rotation is observed during RPT5 disengagement from substrate. The right panel, viewed from a perspective rotated vertically against the left panel, shows that out-of-plane rotation in RPT5 is more prominently amplified in its counterclockwise neighboring ATPases more than its clockwise neighbors. Red arrows mark the center of the AAA ring. b Structural comparison between ED2 (color) and ED1 (grey) in which the large AAA subdomain of RPT5 is used to align the two AAA-ring structures together. A 7° out-of-plane rotation of RPT5 is observed during RPT5 re-association with the substrate. c Schematic illustrates the mechanism for processive substrate translocation that involves synchronization of nucleotide processing in three adjacent ATPases, i.e., ATP binding, ADP release and ATP hydrolysis (left), creating vertical rotations in the ATPases that cooperatively propel the substrate (right)
The aforementioned analysis postulates on a force generation model integrating various structural observations that seem to suggest a high degree of conservation in the operating principle of many AAA-ATPase motors, unfoldases, translocases and disaggregates (Fig. 1.28). To generate processive translocation of a substrate, three adjacent ATPases appear to synchronize their nucleotide processing altogether. When the first binds an ATP, the second releases an ADP and the third hydrolyzes an ATP (Fig. 1.28). ATP binding and ADP release trigger a hinge-like rotation of the AAA domain in opposite directions. Substrate disengagement from the second ATPase provides its AAA domain with certain degrees of freedom to amplify the inter-domain hinge-like rotations in both the first and second ATPases into a collective power-stroke-like motion. The largest out-of-plane rotation (~30°) occurs in the substrate-disengaging ATPase. The power-stroke-like conformational change facilitates ATP hydrolysis in the counterclockwise neighboring ATPase, by repositioning the arginine fingers coordinating the neighboring ATP (Zhang and Wigley 2008).
Coordinated ATP Hydrolysis Around the ATPase Ring
Because most substrate-bound AAA+ ATPase complexes were solved in only single conformation per biochemical condition at high resolution, the sequential model of coordinated ATP hydrolysis around the ATPase ring remains largely hypothetical (Cooney et al. 2019; Puchades et al. 2017; Puchades et al. 2019; Monroe et al. 2017; Ripstein et al. 2017). In few studies where several coexisting conformations were obtained, there was often lack of unambiguous intrinsic features labeling the time sequence of those conformations (de la Pena et al. 2018; Majumder et al. 2019; Fei et al. 2020; Ripstein et al. 2020). Interestingly, two hypothetical interpretations of very similar cryo-EM structures for the ClpXP complex, one proposing a strictly sequential model and the other arguing a probabilistic model, illustrates the limit and uncertainty of inferring biochemical mechanism solely from fewer structure snapshots that are lack of time labels or insufficiently represented in intermediate states necessary to substantiate the hypothesized mechanism (Tsai and Hill 2020; Fei et al. 2020; Ripstein et al. 2020). This controversy underscores the importance of obtaining cryo-EM structures that contain features labeling the temporal sequence, as well as key intermediates unambiguously distinguishing between contradicting or alternative mechanistic models. Breaking such interpretive limits or reducing inference ambiguity might require more structural snapshots along the path of conformational changes. For instance, the seven cryo-EM structures of the substrate-bound human 26S proteasome obtained under a common buffer condition contain inherent features of ubiquitin and substrate densities verifying the time sequence of the corresponding states along the path of chemical reactions (Table 1.2) (Dong et al. 2019; Zhang and Mao 2020). These structures establish themselves as a spatiotemporal continuum and provide direct evidence for sequential ATP hydrolysis around the proteasomal ATPase ring with a mixture of Modes 1, 2 and 3 (Dong et al. 2019). A rigorously sequential Mode-3 hydrolysis around the ring still awaits further evidence.
It is anticipated that the proteasome ATPase motor and other similar systems should be versatile enough to allow for the coexistence of multiple pathways of coordinated ATP hydrolysis. In other words, a rigorously sequential Mode-3 hydrolysis does not necessarily exclude the possible occurrences of ATP hydrolysis that are less sequential or more stochastic. Indeed, recent cryo-EM structures of the yeast substrate-bound Cdc48-Ufd1-Npl4 complex revealed a nearly planar D1 ATPase ring and a spiral-shaped D2 ATPase ring in Mode 2 (Twomey et al. 2019). However, the use of ADP/BeFx resulted in a spiral D1 ring with the D2 ring turning into Mode 3. Similarly, cryo-EM structures of two FtsH-like mitochondrial proteases, the engineered soluble Yme1 and AFG3L2 complexes, revealed conformations of substrate-bound ATPase ring highly resembling Modes 3 and 2 of the 26S proteasome, respectively (Puchades et al. 2017; Puchades et al. 2019). Thus, the fate of the ATP hydrolysis pathway in the hexameric ATPase ring is energetically dependent of its interactions with substrates and regulatory proteins, such as chaperones, shuttle proteins or cofactors.
Summary of Mechanism of the Proteasomal AAA-ATPase Motor
The proteasomal AAA-ATPase motor distinguishes itself from other homohexameric ATPase complexes in several unique ways. First, its pre-engaged conformation is poised to start highly coordinated ATP hydrolysis from RPT6. This role of RPT6 is already encoded in the proteasome structure in the resting state. Second, the mode of coordinated ATP hydrolysis is tightly timed to switch for regulating the necessary intermediate steps of substrate processing. Third, the conformation of the ATPase ring during processive substrate translocation favors fewer major conformational states like ED1 and ED2 other than six major states (Fig. 1.26) that are expected for a rigorous sequential model, indicating a broken symmetry for the kinetic role of each RPT in substrate translocation (Dong et al. 2019; de la Pena et al. 2018). This also prompts the possibility that the sequential hydrolysis model may neither necessarily nor sufficiently account for all translocation activities. To date, the differential appearances of ubiquitin-substrate densities intrinsic to these structures are the only pieces of evidence for labeling the time sequence of the corresponding conformations along the pathway of chemical reactions (Dong et al. 2019). At least three modes of coordinated ATP hydrolysis have been observed to regulate intermediate steps of substrate processing in the functional proteasome (Dong et al. 2019). Remarkably, each mode of coordinated ATP hydrolysis was comparably observed in separate structural snapshots of various AAA+ ATPases under specific biochemical conditions by independent studies (Glynn et al. 2009; Puchades et al. 2019; Cooney et al. 2019; Twomey et al. 2019; Puchades et al. 2017; de la Pena et al. 2018; Yu et al. 2018; Gao et al. 2019; Ripstein et al. 2020; Deville et al. 2019; Puchades et al. 2020; Zhang and Mao 2020). These studies collectively suggest highly conserved dynamic patterns in the structure-function relationships of AAA+ ATPase hexamers that appear to coexist in the proteasome (Dong et al. 2019; Zhang and Mao 2020).
Mechanism of Substrate Degradation in the CP
Structural Mechanism of Substrate Proteolysis
The α- and β-type subunits of the proteasome are typical members in the superfamily of N-terminal nucleophile (Ntn) hydrolases (Brannigan et al. 1995; Dodson and Wlodawer 1998). Consistent with their sequence similarity, the α- and β-subunits exhibit homologous folds (Lowe et al. 1995; Groll et al. 1997). The N-terminal regions (~ 35 residues) of the α-subunits partly fold into a helix (H0) blocking a groove in the β-stranded sandwich. Due to lack of H0, this cleft is left open in the β-subunits, is referred to as S1 (“specificity”) pocket and harbors the active site at Thr1 (Groll et al. 1997). A catalytic triad formed at the N-terminal threonine nucleophile constitutes the active site and is activated upon CP maturation by proteolytic removal of a proximal propeptide occluding the active site (Chen and Hochstrasser 1996; Schmidtke et al. 1996; Seemuller et al. 1996; Huber et al. 2016; Li et al. 2016b). The sidechain Oγ of the N-terminal residue Thr1 of the β-subunits provides the catalytic nucleophile attack of the carbonyl carbon in a peptide bond (Fig. 1.29) (Seemuller et al. 1995a; Lowe et al. 1995). Similar to the proposed hydrolytic mechanism in penicillin acylase (Duggleby et al. 1995), the amino group of Thr1 might be the proton acceptor when Thr1 Oγ adds to an electrophilic entre (Lupas et al. 1995; Seemuller et al. 1995a; Lowe et al. 1995; Groll et al. 1997). Crystal structure of the human CP at 1.8 Å identified a chloride ion, which was previously assigned as catalytic water (NUK), in all active sites, which might aid the nucleophilic attack (Fig. 1.29) (Schrader et al. 2016). The properties of the S1 pocket with respect to substrates are also regulated by other residues of the same subunit and of adjacent subunits β2, β3, and β6, respectively (Groll et al. 1997). Conserved residues essential for the catalytic activity at Thr1 are Glu17 and Lys33 (Seemuller et al. 1995a; Lowe et al. 1995; Arendt and Hochstrasser 1997; Heinemeyer et al. 1997; Seemuller et al. 1996). Other conserved residues, including Ser129, Ser169, and Asp166, seem to be required for structural integrity (Lowe et al. 1995) or β-precursor processing (Seemuller et al. 1995b).
The proteolytically active sites in the 1.8-Å crystal structure of the human CP (PDB ID: 5LE5) (Schrader et al. 2016). a Overview of the proteolytically active site of the β-type subunit of the human CP. Thr1 residues in the β1, β2, and β5 subunits are highlighted as sphere representations, whereas the overall CP is rendered as transparent surface. b–d The close-up view of the residues essential for catalysis around the Thr1 residues in β1 (panel b), β2 (panel c), and β5 (panel d) subunits are shown as stick representations. The chloride ion and water molecules are rendered as green and red spheres, respectively. The distances of the closest water molecules to the side-chain hydroxyl oxygen of Thr1 residue are marked in unit of angstrom
Three β-subunits, β1, β2, and β5, in each β-ring of the eukaryotic CP are proteolytically active. Mutating the Thr1 residues of any of the three β-subunits can disrupt the peptidase activities of the CP (Dick et al. 1998). Similar to classic proteases, the binding pockets are formed at the active sites by specific inter-subunit interactions of the catalytic subunit with its neighboring β-subunit (Borissenko and Groll 2007). The proteasome should not be understood as a simple collection of functionally independent, separated proteases or enzymes but an integral, coherent holoenzyme with multifunctional activities allosterically coupled. Its distributed catalytic activities are spatiotemporally coordinated, and only work properly when the CP is fully assembled and activated. Cleavage of a substrate occurs favorably at the peptide bond located at C-terminal of the P1 residue that is acidic, basic, or hydrophobic (aromatic). Thus, the proteasome displays caspase-like, trypsin-like, and chymotrypsin-like peptide hydrolytic activities (Arendt and Hochstrasser 1997; Voges et al. 1999). These types of activity have been thought to associate with specific β1, β2, and β5 subunits, respectively (Wilk and Orlowski 1983; Heinemeyer et al. 1997; Orlowski 1990; Heinemeyer et al. 1991, 1993; Hilt et al. 1993; Enenkel et al. 1994; Dick et al. 1998). Although the three types of active sites exhibit different but weak preference for substrate sequences, they do not have definite sequence specificity and can cleave peptide bonds at arbitrary positions in an unfolded substrate (Kisselev and Goldberg 2001). Multiple catalytic sites with weak substrate preference offer a unique advantage in concerted processive degradation of diverse cellular proteins by the proteasome (Dick et al. 1991; Bogyo et al. 1998; Wenzel et al. 1994; Ehring et al. 1996; Dolenc et al. 1998; Nussbaum et al. 1998; Arendt and Hochstrasser 1997; Heinemeyer et al. 1997; Groll et al. 1999; Kisselev et al. 1999).
The propeptides of immature β-subunits may play a role in promoting the CP assembly (Chen and Hochstrasser 1996; Mayr et al. 1998; Zuhl et al. 1997). In the newly assembled CP, the propeptides are covalently linked to and bind the Thr1 catalytic sites of the same β-subunits for intramolecular autolysis (Schmidtke et al. 1996; Ditzel et al. 1998). Gly(-1) in the propeptide region is critical for intramolecular processing of the precursor propeptides to activate the β-subunits (Seemuller et al. 1996; Ditzel et al. 1998). Because the catalytic sites also comprise residues of adjacent β-subunits, autolytic activation requires the CP assembly at least in part (Groll et al. 1997). Autolyzed β-subunits (β1, β2, and β5) that are activated may also process the propeptides of other β-subunit precursors (Schmidtke et al. 1996; Ditzel et al. 1998). The propeptides (residues -8 to 1) of the partially processed inactive β-subunits (β3, β6, and β7) are still covalently linked to and shield their Thr1 sites to prevent substrate binding and to inhibit their catalytic activities.
Proteolytic Products by the Proteasome
The CP is thought to degrade substrates in a processive fashion mostly without release of partially degraded intermediates (Dick et al. 1991; Nussbaum et al. 1998; Akopian et al. 1997), although partial degradation by the 26S proteasome has been observed in the activation of many cellular proteins and is likely mediated by the recognition of degradation-stop signal (Saeki et al. 2009a; Zhang et al. 2013; Sandstrom et al. 2019; Chui et al. 2019). The proteolytic chamber efficiently traps unfolded substrates and degrades them into short polypeptides below a certain length limit. The length distribution of released peptide products is broad and ranges from 4 to 25 residues with a length average of 7 to 9 residues (Wenzel et al. 1994; Ehring et al. 1996; Nussbaum et al. 1998; Kisselev et al. 1998). How the CP regulates the product length of degraded substrates has remained enigmatic. One early reasoning attributes to the distance between the active sites that can potentially act in concert (Wenzel et al. 1994). A diffusion-controlled mechanism has been hypothesized (Wenzel et al. 1994; Voges et al. 1999). In the prokaryotic CP, the distance of 2.8 nm between adjacent active sites corresponds to the length of an octa- or nonapeptide in extended conformations (Lowe et al. 1995). However, quantitative analysis of the product length showed a size variation that cannot be reconciled with a solely geometry-based ruler model (Dolenc et al. 1998; Nussbaum et al. 1998; Kisselev et al. 1998). Moreover, these models did not explain why the prokaryotic and eukaryotic proteasomes, which include 14 and 6 active sites, respectively, generate peptides with similar length distributions (Dolenc et al. 1998; Nussbaum et al. 1998; Kisselev et al. 1998).
The possible mechanism of product length regulation by the CP became more comprehensible when more than a dozen non-catalytic substrate-binding sites inside the CP chamber had been observed in the cryo-EM reconstructions of the substrate-engaged human proteasome (Table 1.3) (Dong et al. 2019). Strikingly, all these substrate-binding sites are heavily decorated with aromatic residues, which are also the hallmark of substrate-interacting sites inside the OB and AAA channel. These sites seem to generally stabilize an unfolded peptide segment with a length ranging from 3 to 10 residues and are located in proximity to the three catalytic sites in β1, β2, and β5 (Dong et al. 2019). Their distances to the catalytic sites range from 4 to 25 residues in length, well consistent with the experimental observation of the product length variation. These auxiliary non-catalytic substrate-binding sites may anchor the unstructured substrates during their proteolysis, prohibit refolding of the substrates and prevent the short peptide products from further hydrolytic breakdown. In strong support of this mechanism, long peptides have been observed to degrade faster than short peptides (Dolenc et al. 1998) and NMR studies have shown that the interior of the CP stabilizes unstructured conformations of translocated substrates, which prevents refolding of substrates inside the CP that would sterically hinder their degradation (Ruschak et al. 2010).
Actions of the CP Inhibitors
Structural studies using enzyme inhibitor and site-directed mutagenesis have provided insights into the catalytic mechanism of the CP (Groll and Huber 2004). A crystal structure of the CP inhibited by the peptide aldehyde Ac-Leu-Leu-nLeu-al (ALLN) reveals a hemiacetyl bond with the N-terminal threonine (Thr1) hydroxyl groups of β-subunits (Groll and Huber 2004; Vinitsky et al. 1992; Rock et al. 1994; Figueiredo-Pereira et al. 1994; Vinitsky et al. 1994; Harding et al. 1995). Indeed, mutation of Thr1 to serine retains proteolytic activity, whereas mutation to alanine completely abolishes the proteolytic activity (Groll and Huber 2004). Covalent attachment of a Streptomyces metabolite lactacystin to Thr1 irreversibly inhibits their proteolytic activity (Fenteany et al. 1995). Lactacystin hydrolysis produces clasto-lactacystin beta-lactone that covalently reacts with the active sites (Dick et al. 1996). Other covalent inhibitors have also been investigated, including peptidyl vinyl sulfones (Bogyo et al. 1997b, 1998), dipeptidyl boronic acids (Mc Cormack et al. 1997; Adams et al. 1998) and epoxyketones (Groll and Huber 2004). Most of the covalent inhibitors share a common mechanism of action via reaction with Thr1 Oγ in the β-subunits (Mc Cormack et al. 1997). These CP inhibitors have been extensively used for dissecting the cellular functions of the proteasomes (Bogyo et al. 1997a; Lee and Goldberg 1998; Fenteany and Schreiber 1998).
Since the CP inhibition is an approved approach for cancer therapy, numerous crystal structures of the inhibitor-bound CP complexes have been determined (Huber and Groll 2012; Schrader et al. 2016). The crystal structure of the immunoproteasome CP demonstrates that the active site architecture of the inducible subtype β-subunits (β1i, β2i, and β5i) differs from that of the constitutive CP (Huber et al. 2012). The variation in specificity largely occurs through changes in the S1 pocket and explains why the inhibitor PR-957 specifically binds the β5i subunit. Since the immunoproteasome represents a minor fraction of the proteasome population and is found predominantly in cells involved in the immune response, it is expected that treatment with immunoproteasome-specific inhibitors could result in reduced toxicity (Huber and Groll 2012). Higher resolution (1.9 Å) crystal structure of the human CP in complex with the inhibitor Oprozomib revealed a ring-shaped electron density in the inhibitor-Thr1 conjugate, which was modeled as a heptagonal 1,4-oxazepane ring structure with the C6-methyl group pointed to the inner, solvent-inaccessible side of the ring (Schrader et al. 2016). Thus, the inhibition reaction is mediated via nucleophile attack by the N-terminal amine of the epoxide β carbon (Schrader et al. 2016).
It is also of great clinical merit in developing inhibitors against the proteasomes of infectious pathogens, such as Mycobacterium tuberculosis that causes tuberculosis (Lin et al. 2009), the malaria parasite Plasmodium falciparum (Li et al. 2016a) and the kinetoplastid parasites Trypanosoma cruzi, Leishmania donovani and Trypanosoma brucei (Khare et al. 2016). Interestingly, binding of oxathiazole-2-one inhibitors induces conformational changes of the M. tuberculosis CP, whereas no comparable conformational changes are induced in the human proteasome (Lin et al. 2009). The comparative studies explain why these compounds exhibit specificity for the pathogen proteasome, which provides an important mechanistic guidance for structure-based design of other proteasome inhibitors targeting parasite pathogens (Li et al. 2016a; Khare et al. 2016). Cryo-EM structure of the inhibitor-bound P. falciparum proteasome CP provides information of the active-site conformation that can be used for further refinement of inhibitor design (Li et al. 2016a). Furthermore, NMR studies have demonstrated that CP inhibition can also be achieved by binding near the interface between α and β subunits instead of binding directly to the active sites (Sprangers et al. 2008). The novel structural determination tools in cryo-EM and NMR are expected to offer new approaches of developing proteasome inhibitors (Renaud et al. 2018; Scapin et al. 2018).
Proteasome in Health and Disease
Cancer
The proteasome plays a central role in regulating cellular proteostasis, and is recognized as an important regulator of carcinogenesis (Voutsadakis 2017). The proteasome controls the fate of many short-lived proteins regulating cell cycle and tumor suppressors promoting cycle progression, such as cyclin B1, p21, p27 and p53 (Dietrich et al. 1996; Machiels et al. 1997; Adams et al. 1999; Wu et al. 2000; Shah et al. 2001). Dysregulation of these regulatory proteins is associated with malignancies and carcinogenesis. Most cancer cells survive on a considerable level of proteasome activity and are more susceptible to inhibition of proteasome function than are normal cells (Dulic et al. 1994; Pagano et al. 1995; King et al. 1996; Delic et al. 1998; Orlowski et al. 1998). Many cancer types show aberrant functions in their UPS (Hoeller and Dikic 2009; Chen and Madura 2005). A decreased activity of the proteasome has been found in cancer stem cells compared to the rest of cancer cells (Voutsadakis 2017). Proteasome inhibition disrupts cellular protein homeostasis and cell cycle, and potentially activates programmed cell death and apoptosis. The growth of multiple myeloma cell lines is inhibited by several proteasome inhibitors and exhibits anti-inflammatory and immunosuppressive effects (Muchamuel et al. 2009; Basler et al. 2010; Voorhees and Orlowski 2006). Treatment with proteasome inhibitors has been proven beneficiary for patients with multiple myeloma and mantle cell lymphoma in human clinical trials (Orlowski et al. 2002; Jagannath et al. 2004; Roeten et al. 2018). Upstream of the ubiquitin recognition by the proteasome, many E3 ligases and the ATPase p97/VCP that contribute to substrate selectivity for the proteasome are also implicated in cancers (Tang and Xia 2016; Chapman et al. 2011; Kimonis et al. 2008), and have been considered as potential anti-cancer drug targets due to their crucial roles in proteostasis, protein quality control and cell viability (Anderson et al. 2015; Vekaria et al. 2016; Magnaghi et al. 2013; Skrott et al. 2017).
In clinical trials, the proteasome inhibitor Bortezomib or Velcade (initially called PS-341) exhibited inhibitory efficacy specifically toward hematologic malignancies (Orlowski et al. 2002; Jagannath et al. 2004). Bortezomib is a reversible peptide boronate inhibitor, which preferentially inhibits the chymotrypsin-like activity of the β5 subunit (Huber and Groll 2012). Bortezomib was approved by the US Food and Drug Administration (FDA) for the treatment of refractory multiple myeloma and mantle cell lymphoma (Ruggeri et al. 2009). It also exhibits certain efficacy in treatment of non-small cell lung cancer and pancreatic cancer (Frankland-Searby and Bhaumik 2012). However, Bortezomib treatment gives rise to the development of drug resistance and causes adverse side effects, including asthenia, pain, peripheral neuropathy, disorders, thrombocytopenia, and gastrointestinal, cardiac and pulmonary disorders (Frankland-Searby and Bhaumik 2012).
Several next-generation proteasome inhibitors have been developed for the treatment of hematologic malignancies and solid tumors (Roeten et al. 2018). Some of these inhibitors also target the chymotrypsin-like activity of the β5 subunit, and exhibit increased chemical stability and proteasome-binding affinities, as well as altered toxicities in clinical trials (Huber and Groll 2012). The US FDA approved the proteasome inhibitor Carfilzomib in 2012 for the treatment of patients with multiple myeloma. In contrast to Bortezomib, Carfilzomib is an irreversible peptide epoxyketone inhibitor of the CP (Wang et al. 2013). However, Carfilzomib has shown efficacy in multiple myeloma patients who developed drug resistance against Bortezomib (Wang et al. 2013).
In addition to the inhibitors targeting the CP, a bis-benzylidine piperidone RA190 and its derivatives like RA183 were developed as covalent inhibitors of RPN13 for the treatment of ovarian cancer (Anchoori et al. 2013, 2018; Kisselev 2013; Lu et al. 2017a) and multiple myeloma (Song et al. 2016, 2019; Trader et al. 2015). NMR structural studies found that RA190-binding site on RPN13 PRU domain coincides with the interaction site of RPN13 with a proline-rich C-terminal extension of RPN2 (Lu et al. 2017a). PA190 is found to also directly bind and inactivate UCH37, causing accumulation of substrates at the proteasome. Thus, it has been proposed that RA190 targets both RPN13 and UCH37 in a parallel fashion to effectively block substrate degradation (Lu et al. 2017a).
Aging
Progressive aggregation of damaged or misfolded proteins is a hallmark of aging cells, and is associated with decline in cellular proteostasis function and UPS impairment, which is also implicated in aging-related disease like Alzheimer’s disease (AD) and dementia (Vilchez et al. 2014; Hegde et al. 2019; Fischer et al. 2009; Saez and Vilchez 2014). The ability of the proteasome in maintaining proteostatsis is generally challenged and prone to decline in aging cells, tissues and organisms, such as human epidermal cells and lymphocytes, bovine eye lens and rat muscle, lung, liver, kidney, hippocampus, spinal cord, cerebral cortex and heart (Baraibar et al. 2012; Saez and Vilchez 2014). Age-related proteasome dysregulation may occur at multiple levels, including suppressed expression of proteasome subunits (Lee et al. 1999), reduced proteolytic activity caused by oxidative damage (Bulteau et al. 2001; Wang et al. 2010), and proteasome disassembly (Wang et al. 2010; Vernace et al. 2007; Bajorek et al. 2003). Part of these is also reflected in altered function in ubiquitin ligases, such as cytoplasmic E3 ligase mahogunin (MGRN1) that is found instead in nucleus in aging hippocampal neurons (Benvegnu et al. 2017), and the reduced level of UBE3A in the hippocampus of elderly rats (Fletcher et al. 2014).
Aggregated proteins might form non-productive, stalled complex with the proteasome, deplete the intracellular proteasome reservoir and thus reduce their overall cellular activity (Grune et al. 2004). When the constitutive β5 subunit was substituted with the thymus-specific subtype β5t subunit of reduced chymotrypsin-like activity, the transgenic mice developed signs of early aging and suffered a much shorter lifespan (Tomaru et al. 2012). Consistently, several studies on long-lived humans (centenarians) and animals, such as the exceptionally long-lived naked mole rat and a long-lived bat species and an exceptionally long-lived invertebrate called giant clam, have found correlates with increased proteasome activity and capacity (Chondrogianni et al. 2000; Perez et al. 2009; Salmon et al. 2009; Ungvari et al. 2013). These data suggest that the overall level of proteasome activity may be correlated with the age of an organism. This hypothesis has been further supported by genetic studies in several model organisms (Chen et al. 2006; Tonoki et al. 2009; Kruegel et al. 2011).
Neurodegenerative Diseases
Aggregation of misfolded proteins compromises cellular proteostasis and is characteristic of neurodegenerative diseases, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), Amyotrophic lateral sclerosis (ALS) and spongiform encephalopathies (Selkoe 2003; Rubinsztein 2006; Brettschneider et al. 2015). Maintaining appropriate proteostasis in neurons is crucial due to their complex organization, function and prolonged lifespan. Unchecked aggregation of misfolded proteins can impair normal cellular function in the cytoplasm, nucleus or extracellular space. More importantly, neurons are unable to dilute aggregate loads through cell division (Tai and Schuman 2008). These underscore the critical function of the UPS in neuronal synapses. For example, synaptic protein homeostasis, plasticity and long-term memory formation rely on the proteome remodeling that is tightly regulated by the UPS (Fonseca et al. 2006; Tai and Schuman 2008; Aso et al. 2012; Djakovic et al. 2012). The membrane-anchored proteasome modulates neuronal function in the mammalian nervous system, degrades intracellular proteins and releases the polypeptide products into the synaptic cleft, where they trigger neuronal signaling via postsynaptic N-methyl-D-aspartate receptor for synaptic regulation (Ramachandran and Margolis 2017).
Neurodegenerative diseases are mostly associated with aging, and share many traits in proteostasis, such as impaired UPS function and reduced proteasome activity (Vilchez et al. 2014; Hegde et al. 2019; Fischer et al. 2009; Saez and Vilchez 2014; Keller et al. 2000; McNaught et al. 2002; Ciechanover and Brundin 2003; Rubinsztein 2006; Ortega et al. 2007; Ortega and Lucas 2014). Aggregation of misfolded proteins causes loss of proteostasis and is commonly accompanied with progressive death of neurons (Selkoe 2003; Rubinsztein 2006; Brettschneider et al. 2015). Brain region-specific proteasome inhibition is implicated in the neuropathology and is a clinical hallmark of neurodegenerative diseases (McNaught et al. 2002, 2004; Bedford et al. 2008; Li et al. 2010). In the mice model of AD with mutant amyloid precursor protein (APP) transgene (Tg2576), Aβ1-42 seems to inhibit proteasome activity and DUBs and enhances amyloid and tau accumulation (Almeida et al. 2006; Oh et al. 2005; Tseng et al. 2008), likely by stalling the proteasome in the Aβ aggregation body (Guo et al. 2018). Interestingly, a higher level of immunoproteasome expression was observed in the brain of AD patients compared to the brains of the non-demented elderly (Mishto et al. 2006). Going forward beyond the “Aβ hypothesis” of AD (Morris et al. 2018; Mullane and Williams 2019), it remains to be understood how the proteasome and UPS are related to other factors contributing to neurodegenerative diseases, such as inflammation and insulin resistance in the brain.
Secretory proteins and integral membrane proteins are synthesized and enter the ER lumen for proper protein folding and post-translational modifications. Misfolded proteins in the ER are removed by the pathway of endoplasmic reticulum-associated protein degradation (ERAD), which prevents neurodegenerative diseases (Wu and Rapoport 2018). Impaired ERAD and ER stress are associated with polyglutamine (polyQ) toxicity commonly found in several neurodegenerative disorders (Duennwald and Lindquist 2008; Remondelli and Renna 2017; Smith and Mallucci 2016). There are three distinct ERAD pathways, ERAD-L, ERAD-M, and ERAD-C, in which the misfolded protein domain is localized in the ER lumen, within the membrane, or on the cytosolic side of the ER membrane, respectively (Huyer et al. 2004; Vashist and Ng 2004; Carvalho et al. 2006). Another pathway is used to remove misfolded protein from the inner nuclear membrane (Foresti et al. 2014; Khmelinskii et al. 2014). Through ubiquitylation, the p97/Cdc48 ATPase extracts the substrates from the membrane and delivers them to the proteasome for degradation (Bays et al. 2001; Braun et al. 2002; Jarosch et al. 2002; Rabinovich et al. 2002). Proteasome inhibition impairs ERAD and allows misfolded proteins to aggregate in the ER, which activates the unfolded protein response (UPR) (Tsai and Weissman 2010). UPR regulates the gene expression controlling protein folding and ERAD to restore ER homeostasis (Travers et al. 2000). However, prolonged ER stress that is out of control by UPR activation eventually leads to cell death (Travers et al. 2000). Besides neurodegenerative diseases, many other physiological conditions, such as hypoxia, glucose deprivation, oxidative stress and certain mutations in proteins, can also cause aggregation of misfolded proteins in the ER and trigger UPR activation (Tsai and Weissman 2010).
Immune Diseases
The proteasome is extensively involved in the regulation of the immune system, including antigen presentation, NF-κB pathway, and NLRP1 inflammasome activation. The proteasome mediates antigenic protein degradation into short peptides that are presented on the MHC-I complex (Ferrington and Gregerson 2012). The NF-κB transcription factors (NF-κB and Rel proteins) regulate gene expression in innate and adaptive immunity, inflammation, stress responses, B-cell development and lymphoid organogenesis. NF-κB is a prosurvival pathway and is upregulated in many inflammatory diseases and cancers (Wang et al. 1996). In cancer cells, NF-κB is involved in the expression of the antiapoptotic IAP family of genes as well as BCL-2 prosurvival genes (Wang et al. 1998; Zong et al. 1999; Chen et al. 2000). The proteasome mediates degradation of regulatory elements for transcriptional activation in both canonical and noncanonical pathways of NF-κB activation. In the inactive state, NF-κB/Rel is bound to and sequestered by the inhibitory IκB subunits in the cytoplasm (Baldwin 2001). In the canonical pathway, proinflammmatory cytokines activate the IκB kinase (IKK) complex that phosphorylates IκB and causes IκB ubiquitiylation for proteasome-mediated degradation that activates the NF-κB/RelA complex (Chen et al. 1995; Scherer et al. 1995; Spencer et al. 1999; Winston et al. 1999). In the noncanonical pathway, the NF-κB-inducing kinase activates IKKα that phosphorylates the C-terminal residues of NF-κB2/p100. Phosphorylated NF-κB is ubiquitylated and partially degraded by the proteasome into transcriptionally active NF-κB2/p52.
Mammalian cells use a diverse spectrum of pattern-recognition receptors (PRRs) to monitor cytoplasmic microbial activities and pathogen invasion for signaling innate immune responses. Several PRRs form megadalton multiprotein complexes named the inflammasome upon recognition of their cognate danger signals. The inflammasome recruits and activates caspase-1, which then cleaves and activates inflammatory cytokines and gasdermin D (GSDMD), and triggers inflammatory cell death called pyroptosis (Zhang et al. 2015; Broz and Dixit 2016; Sharif et al. 2019). NLRP1B is one of the inflammasome-forming PRRs and a crucial member in the nucleotide-binding domain leucine-rich repeat (NLR) protein superfamily. Several studies have found that proteasomal degradation of N-terminal domains of the NLRP1B protein releases the C-terminal fragment of the NLRP1B containing a caspase activation and recruitment domain (CARD) domain, which is sufficient to self-assemble, recruit caspase-1 and active the inflammasome (Sandstrom et al. 2019; Chui et al. 2019). The degradation-dependent activation of inflammasome likely underlies its ability to sense any pathogen effectors that can induce proteasomal degradation of NLRP1B, such as IpaH7.8, an E3 ubiquitin ligase secreted by the pathogen Shigella flexneri that ubiquitylates NLRP1B and lead to inflammasome activation in Shigella-infected macrophages (Sandstrom et al. 2019).
Proteasome inhibition can give rise to decreased inflammatory and immune responses, as well as compromised cell migration and adhesion (Frankland-Searby and Bhaumik 2012). These effects are also reflected in the proteasome function in antigen presentation. Lymphopenia is a toxicity effect commonly observed in clinical administration of Bortezomib (Jagannath et al. 2004). Proteasome inhibition by Bortezomib induces suppressive effects on the T cell compartment, potentially leading to T cell apoptosis in monocyte-derived dendritic cells (Nencioni et al. 2006). Activated T cells are more susceptible to proteasome inhibition than resting T cells (Blanco et al. 2011). Proteasome inhibition by Bortezomib can deplete alloreactive T cells in vitro, while preserving immune response against pathogens (Blanco et al. 2011). These observations suggest that proteasome inhibition represents a promising option for the treatment of diseases involving activated T cells. This hypothesis is supported by several preclinical studies in mouse models for autoimmune encepahlomyelitis, arthritis, colitis, graft-versus-host disease, systemic lupus erythematosus, myastenia gravis and allograft rejection (Moran et al. 2012).
Cardiovascular Diseases
The UPS plays an important role in cardiac physiology and disease (Drews and Taegtmeyer 2014; Li and Wang 2011; Pagan et al. 2013; Shukla and Rafiq 2019). Impairment of UPS function may cause a number of cardiac diseases, such as heart failure, cardiomyopathies, hypertrophy, atrophy, ischemia-reperfusion, and atherosclerosis. Increased oxidative damage to proteins, elevated levels of ubiquitylated proteins and proteasome dysfunction have been found in several cardiac diseases, such as hypertrophic, diabetic cardiomyopathies and ischemic heart diseases (Powell et al. 2012; Shukla and Rafiq 2019). Reduced proteasome activity has been linked to myocardial ischemia/reperfusion (I/R) injury (Bulteau et al. 2001; Tian et al. 2012; Li et al. 2011). A transgenic mouse line that overexpresses PA28α in the heart exhibited increased proteasome activity, which protected the animals against I/R injury (Li et al. 2011). In a mouse model of desmin-related cardiomyopathy (DRC), cardiac PA28α overexpression suppressed cardiac hypertrophy and prolonged the animal lifespan (Li et al. 2011). In many forms of cardiomyopathies, such as I/R injury and diabetic cardiomyopathy, inducible immunoproteasome expression is significantly elevated (Cai et al. 2008; Gomes et al. 2006; Powell et al. 2008). Similar to neurological toxicity, proteasome inhibition with Bortezomib seems to be associated with an increase in cardiac dysfunction, such as congestive heart failure or arrythmia (Schlossarek and Carrier 2011). Short-term and low-dose local treatments with proteasome inhibitors have produced contradictive results that await further clarification (Powell et al. 2012). Although both the inhibition and enhancement of proteasome activity can potentially confer cardioprotection under distinct conditions, concerns and controversies remain for the toxicity of impairing the proteasome in long-term therapies (Pagan et al. 2013; Li and Wang 2011; Shukla and Rafiq 2019).
Perspective
In this chapter, I reviewed nearly five decades of studies on the structures and functions of the proteasome. The past five years have seen a major culmination of breakthrough studies in elucidation of the inner working of the proteasome in both human and yeast. Progressive improvements on the biochemical and cryo-EM methods have yielded atomic-level information critical for understanding the functional dynamics of the proteasome. Not only have the conformational states of the proteasome been extensively mapped, but also their correspondence to the intermediate steps of substrate processing and a nearly complete degradation pathway within the proteasome have been reconstructed at the atomic level. A breathtaking picture of the “central dogma” of the proteasome is now emerging and lays an essential foundation for further understanding how the UPS decodes the ubiquitin signals as well as for the development of proteasome-related therapeutics. Looking forward, a major knowledge gap exists between numerous ubiquitylation pathways and the intrinsic working machineries of the proteasome, the understanding of which poses much greater technical challenges due to the transient nature of the interactions of the proteasome with the extrinsic receptors, DUBs, the ubiquitin chain-elongating E4s, and an expanding network of proteasome-related proteins. Another great challenge is how we can best convert what we know about the proteasome into clinical tools and therapeutics for combating human diseases and improving human wellbeing. With further developments in chemical and biological methods, cryo-EM imaging approaches and data science tools, addressing these challenges will soon become possible in years to come.
Future Problems
-
What are the missing key intermediate states necessary for unambiguously defining the detailed mechanisms for the complete cycle of substrate processing in the proteasome? And, how are these intermediate states associated with the proteasome function in vivo?
-
Are there alternative pathways of coordinated ATP hydrolysis in the proteasomal AAA-ATPase motor? How are these hydrolytic pathways regulated at the proteasome level?
-
How do the intrinsic ubiquitin receptors recognize the polyubiquitin chains in the proteasome? What are the conformational dynamics of RPN13 in the proteasome? What are the unidentified intrinsic ubiquitin or UBL receptors in the proteasome? How do they coordinate with one another? What are their functional differences and specificities in recognizing ubiquitin signals and UBL proteins?
-
How do the extrinsic ubiquitin receptors deliver substrates to the proteasome? How do the extrinsic receptors coordinate their ubiquitin recognitions with the intrinsic receptors? What are the other cellular proteins involved in the delivery of substrates by the extrinsic receptors? Are there any other unidentified extrinsic ubiquitin receptors associated the proteasome? What are the differences in substrate selectivity between the intrinsic and extrinsic ubiquitin receptors?
-
How do those non-essential DUBs such as USP14 and UCH37 work with the proteasome? How do their interactions regulate the proteasome structure and function during substrate processing? Are they involved in the process of substrate delivery by the shuttle receptors?
-
How do the proteasome-associated E3 and E4 enzymes process substrates in the proteasome? How do their interactions regulate the proteasome structure and function? Are there also other E2s and E3s associated with the proteasome for substrate processing? How do they work together?
-
How do the proteasome-associated proteins function in the proteasome in the presence of substrates? How do their interactions with the proteasome regulate the proteasome structure and function?
-
How do those post-translational modifications of the proteasome regulate the ways the proteasome processes substrates? What are their cellular functions?
-
What are the assembly pathways of the RP in mammalian cells? How does the dynamic assembly of the RP affect or regulate the proteasome function?
-
Are there inhibitors of the proteasome specifically interacting with the RP instead of the CP? If any, how do these inhibitors contribute to the improvement of human health?
-
What are the exact underlying mechanisms of proteasome inhibitors in vivo? Why is the same proteasome inhibitor beneficiary for certain diseases but detrimental for others?
-
How can we best harvest the proteasomes or UPS for combating neurodegenerative diseases, and of course, other closely related human diseases in general, including cancer and immune diseases?
References
Adams J, Behnke M, Chen S, Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma YT, Plamondon L, Stein RL (1998) Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg Med Chem Lett 8(4):333–338. https://doi.org/10.1016/s0960-894x(98)00029-8
Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, Maas J, Pien CS, Prakash S, Elliott PJ (1999) Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res 59(11):2615–2622
Adrain C, Creagh EM, Cullen SP, Martin SJ (2004) Caspase-dependent inactivation of proteasome function during programmed cell death in Drosophila and man. J Biol Chem 279(35):36923–36930. https://doi.org/10.1074/jbc.M402638200
Ahn JY, Tanahashi N, Akiyama K, Hisamatsu H, Noda C, Tanaka K, Chung CH, Shibmara N, Willy PJ, Mott JD et al (1995) Primary structures of two homologous subunits of PA28, a gamma-interferon-inducible protein activator of the 20S proteasome. FEBS Lett 366(1):37–42. https://doi.org/10.1016/0014-5793(95)00492-r
Aki M, Shimbara N, Takashina M, Akiyama K, Kagawa S, Tamura T, Tanahashi N, Yoshimura T, Tanaka K, Ichihara A (1994) Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J Biochem 115(2):257–269. https://doi.org/10.1093/oxfordjournals.jbchem.a124327
Akopian TN, Kisselev AF, Goldberg AL (1997) Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J Biol Chem 272(3):1791–1798. https://doi.org/10.1074/jbc.272.3.1791
Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko JM, Beck M, Baumeister W, Engel BD (2017) Proteasomes tether to two distinct sites at the nuclear pore complex. Proc Natl Acad Sci U S A 114(52):13726–13731. https://doi.org/10.1073/pnas.1716305114
Alfano C, Faggiano S, Pastore A (2016) The ball and chain of polyubiquitin structures. Trends Biochem Sci 41(4):371–385. https://doi.org/10.1016/j.tibs.2016.01.006
Alfieri C, Chang L, Barford D (2018) Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13. Nature 559(7713):274–278. https://doi.org/10.1038/s41586-018-0281-1
Almeida CG, Takahashi RH, Gouras GK (2006) Beta-amyloid accumulation impairs multivesicular body sorting by inhibiting the ubiquitin-proteasome system. J Neurosci 26(16):4277–4288. https://doi.org/10.1523/JNEUROSCI.5078-05.2006
Ambroggio XI, Rees DC, Deshaies RJ (2004) JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol 2(1):E2. https://doi.org/10.1371/journal.pbio.0020002
Anchoori RK, Jiang R, Peng S, Soong RS, Algethami A, Rudek MA, Anders N, Hung CF, Chen X, Lu X, Kayode O, Dyba M, Walters KJ, Roden RBS (2018) Covalent Rpn13-Binding inhibitors for the treatment of ovarian cancer. ACS Omega 3(9):11917–11929. https://doi.org/10.1021/acsomega.8b01479
Anchoori RK, Karanam B, Peng S, Wang JW, Jiang R, Tanno T, Orlowski RZ, Matsui W, Zhao M, Rudek MA, Hung CF, Chen X, Walters KJ, Roden RB (2013) A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell 24(6):791–805. https://doi.org/10.1016/j.ccr.2013.11.001
Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M (2015) Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell 28(5):653–665. https://doi.org/10.1016/j.ccell.2015.10.002
Arcus V (2002) OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr Opin Struct Biol 12(6):794–801
Arendt CS, Hochstrasser M (1997) Identification of the yeast 20S proteasome catalytic centers and subunit interactions required for active-site formation. Proc Natl Acad Sci U S A 94(14):7156–7161. https://doi.org/10.1073/pnas.94.14.7156
Arrigo AP, Tanaka K, Goldberg AL, Welch WJ (1988) Identity of the 19S ‘prosome’ particle with the large multifunctional protease complex of mammalian cells (the proteasome). Nature 331(6152):192–194. https://doi.org/10.1038/331192a0
Asai M, Tsukamoto O, Minamino T, Asanuma H, Fujita M, Asano Y, Takahama H, Sasaki H, Higo S, Asakura M, Takashima S, Hori M, Kitakaze M (2009) PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. J Mol Cell Cardiol 46(4):452–462. https://doi.org/10.1016/j.yjmcc.2008.11.001
Asano S, Fukuda Y, Beck F, Aufderheide A, Forster F, Danev R, Baumeister W (2015) Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 347(6220):439–442. https://doi.org/10.1126/science.1261197
Aso E, Lomoio S, Lopez-Gonzalez I, Joda L, Carmona M, Fernandez-Yague N, Moreno J, Juves S, Pujol A, Pamplona R, Portero-Otin M, Martin V, Diaz M, Ferrer I (2012) Amyloid generation and dysfunctional immunoproteasome activation with disease progression in animal model of familial Alzheimer’s disease. Brain Pathol 22(5):636–653. https://doi.org/10.1111/j.1750-3639.2011.00560.x
Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, Goldberg AL, Sakata E, Baumeister W, Forster F (2015) Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci U S A 112(28):8626–8631. https://doi.org/10.1073/pnas.1510449112
Bader M, Benjamin S, Wapinski OL, Smith DM, Goldberg AL, Steller H (2011) A conserved F box regulatory complex controls proteasome activity in Drosophila. Cell 145(3):371–382. https://doi.org/10.1016/j.cell.2011.03.021
Baek GH, Kim I, Rao H (2011) The Cdc48 ATPase modulates the interaction between two proteolytic factors Ufd2 and Rad23. Proc Natl Acad Sci U S A 108(33):13558–13563. https://doi.org/10.1073/pnas.1104051108
Baek K, Krist DT, Prabu JR, Hill S, Klugel M, Neumaier LM, von Gronau S, Kleiger G, Schulman BA (2020) NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature 578(7795):461–466. https://doi.org/10.1038/s41586-020-2000-y
Bajorek M, Finley D, Glickman MH (2003) Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol 13(13):1140–1144. https://doi.org/10.1016/s0960-9822(03)00417-2
Baldwin AS (2001) Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest 107(3):241–246. https://doi.org/10.1172/JCI11991
Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, Falconieri V, Deshaies RJ, Milne JL, Huryn D, Arkin M, Subramaniam S (2016) 2.3 A resolution cryo-EM structure of human p 97 and mechanism of allosteric inhibition. Science 351(6275):871–875. https://doi.org/10.1126/science.aad7974
Baraibar MA, Liu L, Ahmed EK, Friguet B (2012) Protein oxidative damage at the crossroads of cellular senescence, aging, and age-related diseases. Oxid Med Cell Longev 2012:919832. https://doi.org/10.1155/2012/919832
Bard JAM, Bashore C, Dong KC, Martin A (2019) The 26S Proteasome utilizes a kinetic gateway to prioritize substrate degradation. Cell177(2):286–298 e215. https://doi.org/10.1016/j.cell.2019.02.031
Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A (2018) Structure and function of the 26S proteasome. Annu Rev Biochem 87:697–724. https://doi.org/10.1146/annurev-biochem-062917-011931
Barthelme D, Chen JZ, Grabenstatter J, Baker TA, Sauer RT (2014) Architecture and assembly of the archaeal Cdc48*20S proteasome. Proc Natl Acad Sci U S A 111(17):E1687–E1694. https://doi.org/10.1073/pnas.1404823111
Barthelme D, Sauer RT (2012) Identification of the Cdc48*20S proteasome as an ancient AAA + proteolytic machine. Science 337(6096):843–846. https://doi.org/10.1126/science.1224352
Barthelme D, Sauer RT (2013) Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proc Natl Acad Sci U S A 110(9):3327–3332. https://doi.org/10.1073/pnas.1300408110
Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, Martin A (2015) Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat Struct Mol Biol 22(9):712–719. https://doi.org/10.1038/nsmb.3075
Basler M, Dajee M, Moll C, Groettrup M, Kirk CJ (2010) Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J Immunol 185(1):634–641. https://doi.org/10.4049/jimmunol.0903182
Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY (2001) HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell 12(12):4114–4128. https://doi.org/10.1091/mbc.12.12.4114
Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C (1996) Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci U S A 93(2):861–866. https://doi.org/10.1073/pnas.93.2.861
Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, Forster F (2012) Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci U S A 109(37):14870–14875. https://doi.org/10.1073/pnas.1213333109
Beck M, Hurt E (2017) The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18(2):73–89. https://doi.org/10.1038/nrm.2016.147
Beckwith R, Estrin E, Worden EJ, Martin A (2013) Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA + unfoldase. Nat Struct Mol Biol 20(10):1164–1172. https://doi.org/10.1038/nsmb.2659
Bedford L, Hay D, Devoy A, Paine S, Powe DG, Seth R, Gray T, Topham I, Fone K, Rezvani N, Mee M, Soane T, Layfield R, Sheppard PW, Ebendal T, Usoskin D, Lowe J, Mayer RJ (2008) Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci 28(33):8189–8198. https://doi.org/10.1523/JNEUROSCI.2218-08.2008
Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL (2003) ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell 11(1):69–78
Benvegnu S, Mateo MI, Palomer E, Jurado-Arjona J, Dotti CG (2017) Aging triggers cytoplasmic depletion and nuclear translocation of the E3 ligase mahogunin: A function for ubiquitin in neuronal survival. Mol Cell 66(3):358–372 e357. https://doi.org/10.1016/j.molcel.2017.04.005
Besche HC, Haas W, Gygi SP, Goldberg AL (2009) Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 48(11):2538–2549. https://doi.org/10.1021/bi802198q
Beskow A, Grimberg KB, Bott LC, Salomons FA, Dantuma NP, Young P (2009) A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J Mol Biol 394(4):732–746. https://doi.org/10.1016/j.jmb.2009.09.050
Biggins S, Ivanovska I, Rose MD (1996) Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J Cell Biol 133(6):1331–1346. https://doi.org/10.1083/jcb.133.6.1331
Bingol B, Schuman EM (2006) Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature 441(7097):1144–1148. https://doi.org/10.1038/nature04769
Blanco B, Sanchez-Abarca LI, Caballero-Velazquez T, Santamaria C, Inoges S, Perez-Simon JA (2011) Depletion of alloreactive T-cells in vitro using the proteasome inhibitor bortezomib preserves the immune response against pathogens. Leuk Res 35(10):1412–1415. https://doi.org/10.1016/j.leukres.2011.05.018
Blickwedehl J, Agarwal M, Seong C, Pandita RK, Melendy T, Sung P, Pandita TK, Bangia N (2008) Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc Natl Acad Sci U S A 105(42):16165–16170. https://doi.org/10.1073/pnas.0803145105
Bochtler M, Ditzel L, Groll M, Huber R (1997) Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc Natl Acad Sci US A 94(12):6070–6074. https://doi.org/10.1073/pnas.94.12.6070
Bodnar NO, Kim KH, Ji Z, Wales TE, Svetlov V, Nudler E, Engen JR, Walz T, Rapoport TA (2018) Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4. Nat Struct Mol Biol 25(7):616–622. https://doi.org/10.1038/s41594-018-0085-x
Bodnar NO, Rapoport TA (2017) Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell 169(4):722–735 e729. https://doi.org/10.1016/j.cell.2017.04.020
Boehringer J, Riedinger C, Paraskevopoulos K, Johnson EO, Lowe ED, Khoudian C, Smith D, Noble ME, Gordon C, Endicott JA (2012) Structural and functional characterization of Rpn12 identifies residues required for Rpn10 proteasome incorporation. Biochem J 448(1):55–65. https://doi.org/10.1042/BJ20120542
Bogyo M, Gaczynska M, Ploegh HL (1997a) Proteasome inhibitors and antigen presentation. Biopolymers 43(4):269–280. https://doi.org/10.1002/(SICI)1097-0282(1997)43:4%3c269:AID-BIP2%3e3.0.CO;2-T
Bogyo M, McMaster JS, Gaczynska M, Tortorella D, Goldberg AL, Ploegh H (1997b) Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc Natl Acad Sci U S A 94(13):6629–6634. https://doi.org/10.1073/pnas.94.13.6629
Bogyo M, Shin S, McMaster JS, Ploegh HL (1998) Substrate binding and sequence preference of the proteasome revealed by active-site-directed affinity probes. Chem Biol 5(6):307–320. https://doi.org/10.1016/s1074-5521(98)90169-7
Bohn S, Beck F, Sakata E, Walzthoeni T, Beck M, Aebersold R, Forster F, Baumeister W, Nickell S (2010) Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci U S A 107(49):20992–20997. https://doi.org/10.1073/pnas.1015530107
Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ (2010) Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic 11(2):210–220. https://doi.org/10.1111/j.1600-0854.2009.01011.x
Book AJ, Gladman NP, Lee SS, Scalf M, Smith LM, Vierstra RD (2010) Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes. J Biol Chem 285(33):25554–25569. https://doi.org/10.1074/jbc.M110.136622
Borissenko L, Groll M (2007) 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem Rev 107(3):687–717. https://doi.org/10.1021/cr0502504
Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL (2001) A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J 20(18):5187–5196. https://doi.org/10.1093/emboj/20.18.5187
Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, Dunn JG, Rouskin S, Inada T, Frost A, Weissman JS (2012) A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151(5):1042–1054. https://doi.org/10.1016/j.cell.2012.10.044
Brannigan JA, Dodson G, Duggleby HJ, Moody PC, Smith JL, Tomchick DR, Murzin AG (1995) A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378(6555):416–419. https://doi.org/10.1038/378416a0
Braten O, Livneh I, Ziv T, Admon A, Kehat I, Caspi LH, Gonen H, Bercovich B, Godzik A, Jahandideh S, Jaroszewski L, Sommer T, Kwon YT, Guharoy M, Tompa P, Ciechanover A (2016) Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc Natl Acad Sci U S A 113(32):E4639–E4647. https://doi.org/10.1073/pnas.1608644113
Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S (2002) Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J 21(4):615–621. https://doi.org/10.1093/emboj/21.4.615
Bremm A, Moniz S, Mader J, Rocha S, Komander D (2014) Cezanne (OTUD7B) regulates HIF-1alpha homeostasis in a proteasome-independent manner. EMBO Rep 15(12):1268–1277. https://doi.org/10.15252/embr.201438850
Brettschneider J, Del Tredici K, Lee VM, Trojanowski JQ (2015) Spreading of pathology in neurodegenerative diseases: a focus on human studies. Nat Rev Neurosci 16(2):109–120. https://doi.org/10.1038/nrn3887
Breusing N, Grune T (2008) Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol Chem 389(3):203–209. https://doi.org/10.1515/BC.2008.029
Brooks P, Fuertes G, Murray RZ, Bose S, Knecht E, Rechsteiner MC, Hendil KB, Tanaka K, Dyson J, Rivett J (2000) Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem J 346(Pt 1):155–161
Broz P, Dixit VM (2016) Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16(7):407–420. https://doi.org/10.1038/nri.2016.58
Buchberger A, Schindelin H, Hanzelmann P (2015) Control of p 97 function by cofactor binding. FEBS Lett 589(19 Pt A):2578–2589. https://doi.org/10.1016/j.febslet.2015.08.028
Budenholzer L, Cheng CL, Li Y, Hochstrasser M (2017) Proteasome structure and assembly. J Mol Biol 429(22):3500–3524. https://doi.org/10.1016/j.jmb.2017.05.027
Buel GR, Chen X, Chari R, O’Neill MJ, Ebelle DL, Jenkins C, Sridharan V, Tarasov SG, Tarasova NI, Andresson T, Walters KJ (2020) Structure of E3 ligase E6AP with a proteasome-binding site provided by substrate receptor hRpn10. Nat Commun 11(1):1291. https://doi.org/10.1038/s41467-020-15073-7
Buetow L, Huang DT (2016) Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol 17(10):626–642. https://doi.org/10.1038/nrm.2016.91
Bug M, Meyer H (2012) Expanding into new markets–VCP/p97 in endocytosis and autophagy. J Struct Biol 179(2):78–82. https://doi.org/10.1016/j.jsb.2012.03.003
Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI (2001) Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem 276(32):30057–30063. https://doi.org/10.1074/jbc.M100142200
Burcoglu J, Zhao L, Enenkel C (2015) Nuclear import of yeast proteasomes. Cells 4(3):387–405. https://doi.org/10.3390/cells4030387
Burgie SE, Bingman CA, Soni AB, Phillips GN Jr (2012) Structural characterization of human Uch37. Proteins 80(2):649–654. https://doi.org/10.1002/prot.23147
Cai ZP, Shen Z, Van Kaer L, Becker LC (2008) Ischemic preconditioning-induced cardioprotection is lost in mice with immunoproteasome subunit low molecular mass polypeptide-2 deficiency. FASEB J 22(12):4248–4257. https://doi.org/10.1096/fj.08-105940
Carvalho P, Goder V, Rapoport TA (2006) Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126(2):361–373. https://doi.org/10.1016/j.cell.2006.05.043
Cascio P, Call M, Petre BM, Walz T, Goldberg AL (2002) Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J 21(11):2636–2645. https://doi.org/10.1093/emboj/21.11.2636
Castaneda CA, Kashyap TR, Nakasone MA, Krueger S, Fushman D (2013) Unique structural, dynamical, and functional properties of k11-linked polyubiquitin chains. Structure 21(7):1168–1181. https://doi.org/10.1016/j.str.2013.04.029
Chang CW, Lee S, Tsai FTF (2017) Structural elements regulating AAA + protein quality control machines. Front Mol Biosci 4:27. https://doi.org/10.3389/fmolb.2017.00027
Chapman E, Fry AN, Kang M (2011) The complexities of p97 function in health and disease. Mol BioSyst 7(3):700–710. https://doi.org/10.1039/c0mb00176g
Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR (2000) How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J 19(15):4111–4122. https://doi.org/10.1093/emboj/19.15.4111
Chen C, Edelstein LC, Gelinas C (2000) The Rel/NF-kappaB family directly activates expression of the apoptosis inhibitor Bcl-x(L). Mol Cell Biol 20(8):2687–2695. https://doi.org/10.1128/mcb.20.8.2687-2695.2000
Chen L, Madura K (2002) Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol 22(13):4902–4913. https://doi.org/10.1128/mcb.22.13.4902-4913.2002
Chen L, Madura K (2005) Increased proteasome activity, ubiquitin-conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Res 65(13):5599–5606. https://doi.org/10.1158/0008-5472.CAN-05-0201
Chen L, Madura K (2014) Yeast importin-alpha (Srp1) performs distinct roles in the import of nuclear proteins and in targeting proteasomes to the nucleus. J Biol Chem 289(46):32339–32352. https://doi.org/10.1074/jbc.M114.582023
Chen L, Romero L, Chuang SM, Tournier V, Joshi KK, Lee JA, Kovvali G, Madura K (2011) Sts1 plays a key role in targeting proteasomes to the nucleus. J Biol Chem 286(4):3104–3118. https://doi.org/10.1074/jbc.M110.135863
Chen P, Hochstrasser M (1995) Biogenesis, structure and function of the yeast 20S proteasome. EMBO J 14(11):2620–2630
Chen P, Hochstrasser M (1996) Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 86(6):961–972. https://doi.org/10.1016/s0092-8674(00)80171-3
Chen Q, Thorpe J, Dohmen JR, Li F, Keller JN (2006) Ump1 extends yeast lifespan and enhances viability during oxidative stress: central role for the proteasome? Free Radic Biol Med 40(1):120–126. https://doi.org/10.1016/j.freeradbiomed.2005.08.048
Chen S, Wu J, Lu Y, Ma YB, Lee BH, Yu Z, Ouyang Q, Finley DJ, Kirschner MW, Mao Y (2016a) Structural basis for dynamic regulation of the human 26S proteasome. Proc Natl Acad Sci U S A 113(46):12991–12996. https://doi.org/10.1073/pnas.1614614113
Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM (2007) Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell 26(6):843–852. https://doi.org/10.1016/j.molcel.2007.05.022
Chen X, Ebelle DL, Wright BJ, Sridharan V, Hooper E, Walters KJ (2019) Structure of hRpn10 bound to UBQLN2 UBL illustrates basis for complementarity between shuttle factors and substrates at the proteasome. J Mol Biol 431(5):939–955. https://doi.org/10.1016/j.jmb.2019.01.021
Chen X, Lee BH, Finley D, Walters KJ (2010) Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol Cell 38(3):404–415. https://doi.org/10.1016/j.molcel.2010.04.019
Chen X, Randles L, Shi K, Tarasov SG, Aihara H, Walters KJ (2016b) Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 reveal distinct binding mechanisms between substrate receptors and shuttle factors of the proteasome. Structure 24(8):1257–1270. https://doi.org/10.1016/j.str.2016.05.018
Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, Maniatis T (1995) Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Dev 9(13):1586–1597. https://doi.org/10.1101/gad.9.13.1586
Chen ZJ, Sun LJ (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell 33(3):275–286. https://doi.org/10.1016/j.molcel.2009.01.014
Chen X, Dorris Z, Shi D, Huang RK, Khant H, Fox T, de Val N, Williams D, Zhang P, Walters KJ (2020) Cryo-EM Reveals Unanchored M1-Ubiquitin Chain Binding at hRpn11 of the 26S Proteasome. Structure. https://doi.org/10.1016/j.str.2020.07.011
Chondrogianni N, Petropoulos I, Franceschi C, Friguet B, Gonos ES (2000) Fibroblast cultures from healthy centenarians have an active proteasome. Exp Gerontol 35(6–7):721–728. https://doi.org/10.1016/s0531-5565(00)00137-6
Christianson JC, Ye Y (2014) Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol 21(4):325–335. https://doi.org/10.1038/nsmb.2793
Chui AJ, Okondo MC, Rao SD, Gai K, Griswold AR, Johnson DC, Ball DP, Taabazuing CY, Orth EL, Vittimberga BA, Bachovchin DA (2019) N-terminal degradation activates the NLRP1B inflammasome. Science 364(6435):82–85. https://doi.org/10.1126/science.aau1208
Ciechanover A (2005) Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol 6(1):79–87. https://doi.org/10.1038/nrm1552
Ciechanover A, Brundin P (2003) The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40(2):427–446. https://doi.org/10.1016/s0896-6273(03)00606-8
Ciechanover A, Elias S, Heller H, Ferber S, Hershko A (1980a) Characterization of the heat-stable polypeptide of the ATP-dependent proteolytic system from reticulocytes. J Biol Chem 255(16):7525–7528
Ciechanover A, Heller H, Elias S, Haas AL, Hershko A (1980b) ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci U S A 77(3):1365–1368. https://doi.org/10.1073/pnas.77.3.1365
Ciechanover A, Kwon YT (2015) Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med 47:e147. https://doi.org/10.1038/emm.2014.117
Ciehanover A, Hod Y, Hershko A (1978) A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun 81(4):1100–1105. https://doi.org/10.1016/0006-291x(78)91249-4
Clague MJ, Urbe S, Komander D (2019) Breaking the chains: deubiquitylating enzyme specificity begets function. Nat Rev Mol Cell Biol 20(6):338–352. https://doi.org/10.1038/s41580-019-0099-1
Cohen-Kaplan V, Livneh I, Avni N, Fabre B, Ziv T, Kwon YT, Ciechanover A (2016) p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc Natl Acad Sci U S A 113(47):E7490–E7499. https://doi.org/10.1073/pnas.1615455113
Collins GA, Goldberg AL (2017) The logic of the 26S proteasome. Cell 169(5):792–806. https://doi.org/10.1016/j.cell.2017.04.023
Collins GA, Goldberg AL (2020) Proteins containing ubiquitin-like (Ubl) domains not only bind to 26S proteasomes but also induce their activation. Proc Natl Acad Sci U S A 117(9):4664–4674. https://doi.org/10.1073/pnas.1915534117
Cook WJ, Jeffrey LC, Carson M, Chen Z, Pickart CM (1992) Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). J Biol Chem 267(23):16467–16471. https://doi.org/10.2210/pdb1aar/pdb
Cooney I, Han H, Stewart MG, Carson RH, Hansen DT, Iwasa JH, Price JC, Hill CP, Shen PS (2019) Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science 365(6452):502–505. https://doi.org/10.1126/science.aax0486
Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298(5593):608–611. https://doi.org/10.1126/science.1075901
Coux O, Tanaka K, Goldberg AL (1996) Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem 65:801–847. https://doi.org/10.1146/annurev.bi.65.070196.004101
Craney A, Rape M (2013) Dynamic regulation of ubiquitin-dependent cell cycle control. Curr Opin Cell Biol 25(6):704–710. https://doi.org/10.1016/j.ceb.2013.07.004
Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127(7):1401–1413. https://doi.org/10.1016/j.cell.2006.09.051
Cruz M, Elenich LA, Smolarek TA, Menon AG, Monaco JJ (1997) DNA sequence, chromosomal localization, and tissue expression of the mouse proteasome subunit lmp10 (Psmb10) gene. Genomics 45(3):618–622. https://doi.org/10.1006/geno.1997.4977
Cui Z, Scruggs SB, Gilda JE, Ping P, Gomes AV (2014) Regulation of cardiac proteasomes by ubiquitination, SUMOylation, and beyond. J Mol Cell Cardiol 71:32–42. https://doi.org/10.1016/j.yjmcc.2013.10.008
da Fonseca PC, He J, Morris EP (2012) Molecular model of the human 26S proteasome. Mol Cell 46(1):54–66. https://doi.org/10.1016/j.molcel.2012.03.026
Dahlmann B, Ruppert T, Kuehn L, Merforth S, Kloetzel PM (2000) Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J Mol Biol 303(5):643–653. https://doi.org/10.1006/jmbi.2000.4185
Dambacher CM, Worden EJ, Herzik MA, Martin A, Lander GC (2016) Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. Elife 5:e13027. https://doi.org/10.7554/eLife.13027
Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, Rush J, Peng J (2011) Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem 286(12):10457–10465. https://doi.org/10.1074/jbc.M110.149633
Dang FW, Chen L, Madura K (2016) Catalytically active proteasomes function predominantly in the cytosol. J Biol Chem 291(36):18765–18777. https://doi.org/10.1074/jbc.M115.712406
Dange T, Smith D, Noy T, Rommel PC, Jurzitza L, Cordero RJ, Legendre A, Finley D, Goldberg AL, Schmidt M (2011) Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J Biol Chem 286(50):42830–42839. https://doi.org/10.1074/jbc.M111.300178
Dantuma NP, Hoppe T (2012) Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends Cell Biol 22(9):483–491. https://doi.org/10.1016/j.tcb.2012.06.003
Darwin KH (2009) Prokaryotic ubiquitin-like protein (Pup), proteasomes and pathogenesis. Nat Rev Microbiol 7(7):485–491. https://doi.org/10.1038/nrmicro2148
Datta AB, Hura GL, Wolberger C (2009) The structure and conformation of Lys63-linked tetraubiquitin. J Mol Biol 392(5):1117–1124. https://doi.org/10.1016/j.jmb.2009.07.090
Davies CW, Paul LN, Kim MI, Das C (2011) Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability. J Mol Biol 413(2):416–429. https://doi.org/10.1016/j.jmb.2011.08.029
Davies JM, Brunger AT, Weis WI (2008) Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure 16(5):715–726. https://doi.org/10.1016/j.str.2008.02.010
Davies KJ (2001) Degradation of oxidized proteins by the 20S proteasome. Biochimie 83(3–4):301–310. https://doi.org/10.1016/s0300-9084(01)01250-0
De La Mota-Peynado A, Lee SY, Pierce BM, Wani P, Singh CR, Roelofs J (2013) The proteasome-associated protein Ecm29 inhibits proteasomal ATPase activity and in vivo protein degradation by the proteasome. J Biol Chem 288(41):29467–29481. https://doi.org/10.1074/jbc.M113.491662
de la Pena AH, Goodall EA, Gates SN, Lander GC, Martin A (2018) Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362(6418). https://doi.org/10.1126/science.aav0725
de Poot SAH, Tian G, Finley D (2017) Meddling with fate: the proteasomal deubiquitinating enzymes. J Mol Biol 429(22):3525–3545. https://doi.org/10.1016/j.jmb.2017.09.015
Delic J, Masdehors P, Omura S, Cosset JM, Dumont J, Binet JL, Magdelenat H (1998) The proteasome inhibitor lactacystin induces apoptosis and sensitizes chemo- and radioresistant human chronic lymphocytic leukaemia lymphocytes to TNF-alpha-initiated apoptosis. Br J Cancer 77(7):1103–1107. https://doi.org/10.1038/bjc.1998.183
Demartino GN, Gillette TG (2007) Proteasomes: machines for all reasons. Cell 129(4):659–662. https://doi.org/10.1016/j.cell.2007.05.007
Demasi M, Silva GM, Netto LE (2003) 20 S proteasome from Saccharomyces cerevisiae is responsive to redox modifications and is S-glutathionylated. J Biol Chem 278(1):679–685. https://doi.org/10.1074/jbc.M209282200
des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y (2015) Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525(7570):491–495. https://doi.org/10.1038/nature14891
Deveraux Q, Ustrell V, Pickart C, Rechsteiner M (1994) A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem 269(10):7059–7061
Deville C, Carroni M, Franke KB, Topf M, Bukau B, Mogk A, Saibil HR (2017) Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci Adv 3(8):e1701726. https://doi.org/10.1126/sciadv.1701726
Deville C, Franke K, Mogk A, Bukau B, Saibil HR (2019) Two-step activation mechanism of the ClpB disaggregase for sequential substrate threading by the main ATPase motor. Cell Rep 27(12):3433–3446 e3434. https://doi.org/10.1016/j.celrep.2019.05.075
Di Fiore PP, Polo S, Hofmann K (2003) When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol 4(6):491–497. https://doi.org/10.1038/nrm1124
Dibble CC, Manning BD (2013) Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol 15(6):555–564. https://doi.org/10.1038/ncb2763
Dick LR, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Stein RL (1996) Mechanistic studies on the inactivation of the proteasome by lactacystin: a central role for clasto-lactacystin beta-lactone. J Biol Chem 271(13):7273–7276. https://doi.org/10.1074/jbc.271.13.7273
Dick LR, Moomaw CR, DeMartino GN, Slaughter CA (1991) Degradation of oxidized insulin B chain by the multiproteinase complex macropain (proteasome). Biochemistry 30(10):2725–2734. https://doi.org/10.1021/bi00224a022
Dick TP, Nussbaum AK, Deeg M, Heinemeyer W, Groll M, Schirle M, Keilholz W, Stevanovic S, Wolf DH, Huber R, Rammensee HG, Schild H (1998) Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J Biol Chem 273(40):25637–25646. https://doi.org/10.1074/jbc.273.40.25637
Dietrich C, Bartsch T, Schanz F, Oesch F, Wieser RJ (1996) p53-dependent cell cycle arrest induced by N-acetyl-L-leucinyl-L-leucinyl-L-norleucinal in platelet-derived growth factor-stimulated human fibroblasts. Proc Natl Acad Sci U S A 93(20):10815–10819. https://doi.org/10.1073/pnas.93.20.10815
Ding Z, Fu Z, Xu C, Wang Y, Wang Y, Li J, Kong L, Chen J, Li N, Zhang R, Cong Y (2017) High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx. Cell Res 27(3):373–385. https://doi.org/10.1038/cr.2017.12
Ding Z, Xu C, Sahu I, Wang Y, Fu Z, Huang M, Wong CCL, Glickman MH, Cong Y (2019) Structural snapshots of 26S proteasome reveal tetraubiquitin-induced conformations. Mol Cell 73(6):1150–1161 e1156. https://doi.org/10.1016/j.molcel.2019.01.018
Ditzel L, Huber R, Mann K, Heinemeyer W, Wolf DH, Groll M (1998) Conformational constraints for protein self-cleavage in the proteasome. J Mol Biol 279(5):1187–1191. https://doi.org/10.1006/jmbi.1998.1818
Djakovic SN, Marquez-Lona EM, Jakawich SK, Wright R, Chu C, Sutton MA, Patrick GN (2012) Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. J Neurosci 32(15):5126–5131. https://doi.org/10.1523/JNEUROSCI.4427-11.2012
Dodson G, Wlodawer A (1998) Catalytic triads and their relatives. Trends Biochem Sci 23(9):347–352. https://doi.org/10.1016/s0968-0004(98)01254-7
Dolenc I, Seemuller E, Baumeister W (1998) Decelerated degradation of short peptides by the 20S proteasome. FEBS Lett 434(3):357–361. https://doi.org/10.1016/s0014-5793(98)01010-2
Dong Y, Zhang S, Wu Z, Li X, Wang WL, Zhu Y, Stoilova-McPhie S, Lu Y, Finley D, Mao Y (2019) Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 565(7737):49–55. https://doi.org/10.1038/s41586-018-0736-4
Drews O, Taegtmeyer H (2014) Targeting the ubiquitin-proteasome system in heart disease: the basis for new therapeutic strategies. Antioxid Redox Signal 21(17):2322–2343. https://doi.org/10.1089/ars.2013.5823
Dubiel W, Pratt G, Ferrell K, Rechsteiner M (1992) Purification of an 11 S regulator of the multicatalytic protease. J Biol Chem 267(31):22369–22377
Duennwald ML, Lindquist S (2008) Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev 22(23):3308–3319. https://doi.org/10.1101/gad.1673408
Duggleby HJ, Tolley SP, Hill CP, Dodson EJ, Dodson G, Moody PC (1995) Penicillin acylase has a single-amino-acid catalytic centre. Nature 373(6511):264–268. https://doi.org/10.1038/373264a0
Dulic V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, Elledge SJ, Reed SI (1994) p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell 76(6):1013–1023. https://doi.org/10.1016/0092-8674(94)90379-4
Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, Vucic D (2010) c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J 29(24):4198–4209. https://doi.org/10.1038/emboj.2010.300
Edbauer D, Haass C (2016) An amyloid-like cascade hypothesis for C9orf72 ALS/FTD. Curr Opin Neurobiol 36:99–106. https://doi.org/10.1016/j.conb.2015.10.009
Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C (2007) Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol 367(1):204–211. https://doi.org/10.1016/j.jmb.2006.12.065
Ehring B, Meyer TH, Eckerskorn C, Lottspeich F, Tampe R (1996) Effects of major-histocompatibility-complex-encoded subunits on the peptidase and proteolytic activities of human 20S proteasomes. Cleavage of proteins and antigenic peptides. Eur J Biochem 235(1–2):404–415. https://doi.org/10.1111/j.1432-1033.1996.00404.x
Eisele MR, Reed RG, Rudack T, Schweitzer A, Beck F, Nagy I, Pfeifer G, Plitzko JM, Baumeister W, Tomko RJ, Jr., Sakata E (2018) Expanded Coverage of the 26S Proteasome conformational landscape reveals mechanisms of peptidase gating. Cell Rep 24(5):1301–1315 e1305. https://doi.org/10.1016/j.celrep.2018.07.004
Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D (2004) Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem 279(26):26817–26822. https://doi.org/10.1074/jbc.M404020200
Elsasser S, Finley D (2005) Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol 7(8):742–749. https://doi.org/10.1038/ncb0805-742
Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol 4(9):725–730. https://doi.org/10.1038/ncb845
Enenkel C, Blobel G, Rexach M (1995) Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem 270(28):16499–16502. https://doi.org/10.1074/jbc.270.28.16499
Enenkel C, Lehmann A, Kloetzel PM (1998) Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J 17(21):6144–6154. https://doi.org/10.1093/emboj/17.21.6144
Enenkel C, Lehmann H, Kipper J, Guckel R, Hilt W, Wolf DH (1994) PRE3, highly homologous to the human major histocompatibility complex-linked LMP2 (RING12) gene, codes for a yeast proteasome subunit necessary for the peptidylglutamyl-peptide hydrolyzing activity. FEBS Lett 341(2–3):193–196. https://doi.org/10.1016/0014-5793(94)80455-9
Erales J, Coffino P (2014) Ubiquitin-independent proteasomal degradation. Biochim Biophys Acta 1843(1):216–221. https://doi.org/10.1016/j.bbamcr.2013.05.008
Erales J, Hoyt MA, Troll F, Coffino P (2012) Functional asymmetries of proteasome translocase pore. J Biol Chem 287(22):18535–18543. https://doi.org/10.1074/jbc.M112.357327
Erpapazoglou Z, Walker O, Haguenauer-Tsapis R (2014) Versatile roles of k63-linked ubiquitin chains in trafficking. Cells 3(4):1027–1088. https://doi.org/10.3390/cells3041027
Erzberger JP, Berger JM (2006) Evolutionary relationships and structural mechanisms of AAA + proteins. Annu Rev Biophys Biomol Struct 35:93–114. https://doi.org/10.1146/annurev.biophys.35.040405.101933
Estrin E, Lopez-Blanco JR, Chacon P, Martin A (2013) Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure 21(9):1624–1635. https://doi.org/10.1016/j.str.2013.06.023
Etlinger JD, Goldberg AL (1977) A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc Natl Acad Sci U S A 74(1):54–58. https://doi.org/10.1073/pnas.74.1.54
Farre JC, Subramani S (2016) Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol 17(9):537–552. https://doi.org/10.1038/nrm.2016.74
Fei X, Bell TA, Jenni S, Stinson BM, Baker TA, Harrison SC, Sauer RT (2020) Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate. Elife 9. 10.7554/eLife.52774
Fenteany G, Schreiber SL (1998) Lactacystin, proteasome function, and cell fate. J Biol Chem 273(15):8545–8548. https://doi.org/10.1074/jbc.273.15.8545
Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL (1995) Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268(5211):726–731. https://doi.org/10.1126/science.7732382
Ferrington DA, Gregerson DS (2012) Immunoproteasomes: structure, function, and antigen presentation. Prog Mol Biol Transl Sci 109:75–112. https://doi.org/10.1016/B978-0-12-397863-9.00003-1
Figueiredo-Pereira ME, Berg KA, Wilk S (1994) A new inhibitor of the chymotrypsin-like activity of the multicatalytic proteinase complex (20S proteasome) induces accumulation of ubiquitin-protein conjugates in a neuronal cell. J Neurochem 63(4):1578–1581. https://doi.org/10.1046/j.1471-4159.1994.63041578.x
Finley D (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78:477–513. https://doi.org/10.1146/annurev.biochem.78.081507.101607
Finley D, Chen X, Walters KJ (2016) Gates, channels, and switches: elements of the proteasome machine. Trends Biochem Sci 41(1):77–93. https://doi.org/10.1016/j.tibs.2015.10.009
Finley D, Prado MA (2019) The proteasome and its network: engineering for adaptability. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a033985
Fischer DF, van Dijk R, van Tijn P, Hobo B, Verhage MC, van der Schors RC, Li KW, van Minnen J, Hol EM, van Leeuwen FW (2009) Long-term proteasome dysfunction in the mouse brain by expression of aberrant ubiquitin. Neurobiol Aging 30(6):847–863. https://doi.org/10.1016/j.neurobiolaging.2008.06.009
Fishbain S, Inobe T, Israeli E, Chavali S, Yu H, Kago G, Babu MM, Matouschek A (2015) Sequence composition of disordered regions fine-tunes protein half-life. Nat Struct Mol Biol 22(3):214–221. https://doi.org/10.1038/nsmb.2958
Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A (2011) Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun 2:192. https://doi.org/10.1038/ncomms1194
Fletcher BR, Hill GS, Long JM, Gallagher M, Shapiro ML, Rapp PR (2014) A fine balance: regulation of hippocampal Arc/Arg3.1 transcription, translation and degradation in a rat model of normal cognitive aging. Neurobiol Learn Mem 115:58–67. https://doi.org/10.1016/j.nlm.2014.08.007
Flick K, Ouni I, Wohlschlegel JA, Capati C, McDonald WH, Yates JR, Kaiser P (2004) Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat Cell Biol 6(7):634–641. https://doi.org/10.1038/ncb1143
Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nagerl UV (2006) A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron 52(2):239–245. https://doi.org/10.1016/j.neuron.2006.08.015
Foresti O, Rodriguez-Vaello V, Funaya C, Carvalho P (2014) Quality control of inner nuclear membrane proteins by the Asi complex. Science 346(6210):751–755. https://doi.org/10.1126/science.1255638
Forster A, Masters EI, Whitby FG, Robinson H, Hill CP (2005) The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell 18(5):589–599. https://doi.org/10.1016/j.molcel.2005.04.016
Forster A, Whitby FG, Hill CP (2003) The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. EMBO J 22(17):4356–4364. https://doi.org/10.1093/emboj/cdg436
Forster F, Unverdorben P, Sledz P, Baumeister W (2013) Unveiling the long-held secrets of the 26S proteasome. Structure 21(9):1551–1562. https://doi.org/10.1016/j.str.2013.08.010
Frank J (2006) Three-dimensional electron microscopy of macromolecular assemblies : visualization of biological molecules in their native state, 2nd edn. Oxford University Press, Oxford, New York
Frankland-Searby S, Bhaumik SR (2012) The 26S proteasome complex: an attractive target for cancer therapy. Biochim Biophys Acta 1825(1):64–76. 10.1016/j.bbcan.2011.10.003
Franz A, Ackermann L, Hoppe T (2016) Ring of change: CDC48/p97 drives protein dynamics at chromatin. Front Genet 7:73. https://doi.org/10.3389/fgene.2016.00073
Freibaum BD, Taylor JP (2017) The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front Mol Neurosci 10:35. https://doi.org/10.3389/fnmol.2017.00035
Frentzel S, Pesold-Hurt B, Seelig A, Kloetzel PM (1994) 20 S proteasomes are assembled via distinct precursor complexes. Processing of LMP2 and LMP7 proproteins takes place in 13–16 S preproteasome complexes. J Mol Biol 236(4):975–981. https://doi.org/10.1016/0022-2836(94)90003-5
Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD (2001) Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J 20(24):7096–7107. https://doi.org/10.1093/emboj/20.24.7096
Fu H, Sadis S, Rubin DM, Glickman M, van Nocker S, Finley D, Vierstra RD (1998) Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J Biol Chem 273(4):1970–1981. https://doi.org/10.1074/jbc.273.4.1970
Fu X, Sokolova V, Webb KJ, Old W, Park S (2018) Ubiquitin-dependent switch during assembly of the proteasomal ATPases mediated by Not4 ubiquitin ligase. Proc Natl Acad Sci U S A 115(52):13246–13251. https://doi.org/10.1073/pnas.1805353115
Fukunaga K, Kudo T, Toh-e A, Tanaka K, Saeki Y (2010) Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochem Biophys Res Commun 396(4):1048–1053. https://doi.org/10.1016/j.bbrc.2010.05.061
Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H (2002) Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc Natl Acad Sci U S A 99(2):745–750. https://doi.org/10.1073/pnas.012585199
Gan J, Leestemaker Y, Sapmaz A, Ovaa H (2019) Highlighting the proteasome: using fluorescence to visualize proteasome activity and distribution. Front Mol Biosci 6:14. https://doi.org/10.3389/fmolb.2019.00014
Gao Y, Cui Y, Fox T, Lin S, Wang H, de Val N, Zhou ZH, Yang W (2019) Structures and operating principles of the replisome. Science 363 (6429). https://doi.org/10.1126/science.aav7003
Gardner RG, Nelson ZW, Gottschling DE (2005) Degradation-mediated protein quality control in the nucleus. Cell 120(6):803–815. https://doi.org/10.1016/j.cell.2005.01.016
Gates SN, Yokom AL, Lin J, Jackrel ME, Rizo AN, Kendsersky NM, Buell CE, Sweeny EA, Mack KL, Chuang E, Torrente MP, Su M, Shorter J, Southworth DR (2017) Ratchet-like polypeptide translocation mechanism of the AAA + disaggregase Hsp104. Science 357(6348):273–279. https://doi.org/10.1126/science.aan1052
Gatica D, Lahiri V, Klionsky DJ (2018) Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20(3):233–242. https://doi.org/10.1038/s41556-018-0037-z
Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, Cosio DM, van Blitterswijk M, Lee WC, Rademakers R, Boylan KB, Dickson DW, Petrucelli L (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 126(6):829–844. https://doi.org/10.1007/s00401-013-1192-8
Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425(6959):737–741. https://doi.org/10.1038/nature02046
Gladman NP, Marshall RS, Lee KH, Vierstra RD (2016) The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis. Plant Cell 28(6):1279–1296. https://doi.org/10.1105/tpc.15.01022
Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94(5):615–623. https://doi.org/10.1016/s0092-8674(00)81603-7
Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT (2009) Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA + protein-unfolding machine. Cell 139(4):744–756. https://doi.org/10.1016/j.cell.2009.09.034
Godderz D, Heinen C, Marchese FP, Kurz T, Acs K, Dantuma NP (2015) Cdc48-independent proteasomal degradation coincides with a reduced need for ubiquitylation. Sci Rep 5:7615. https://doi.org/10.1038/srep07615
Goebl MG, Goetsch L, Byers B (1994) The Ubc3 (Cdc34) ubiquitin-conjugating enzyme is ubiquitinated and phosphorylated in vivo. Mol Cell Biol 14(5):3022–3029. https://doi.org/10.1128/mcb.14.5.3022
Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA (1975) Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci U S A 72(1):11–15. https://doi.org/10.1073/pnas.72.1.11
Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, Jones RC, Thyparambil S, Wang GW, Qiao X, Bardag-Gorce F, Ping P (2006) Mapping the murine cardiac 26S proteasome complexes. Circ Res 99(4):362–371. https://doi.org/10.1161/01.RES.0000237386.98506.f7
Gomez TA, Kolawa N, Gee M, Sweredoski MJ, Deshaies RJ (2011) Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol 9:33. https://doi.org/10.1186/1741-7007-9-33
Gorbea C, Goellner GM, Teter K, Holmes RK, Rechsteiner M (2004) Characterization of mammalian Ecm29, a 26 S proteasome-associated protein that localizes to the nucleus and membrane vesicles. J Biol Chem 279(52):54849–54861. https://doi.org/10.1074/jbc.M410444200
Gorbea C, Pratt G, Ustrell V, Bell R, Sahasrabudhe S, Hughes RE, Rechsteiner M (2010) A protein interaction network for Ecm29 links the 26 S proteasome to molecular motors and endosomal components. J Biol Chem 285(41):31616–31633. https://doi.org/10.1074/jbc.M110.154120
Greene ER, Goodall EA, de la Pena AH, Matyskiela ME, Lander GC, Martin A (2019) Specific lid-base contacts in the 26 s proteasome control the conformational switching required for substrate degradation. Elife 8. https://doi.org/10.7554/elife.49806
Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D (2000) A gated channel into the proteasome core particle. Nat Struct Biol 7(11):1062–1067. https://doi.org/10.1038/80992
Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R (1997) Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386(6624):463–471. https://doi.org/10.1038/386463a0
Groll M, Heinemeyer W, Jager S, Ullrich T, Bochtler M, Wolf DH, Huber R (1999) The catalytic sites of 20S proteasomes and their role in subunit maturation: a mutational and crystallographic study. Proc Natl Acad Sci U S A 96(20):10976–10983. https://doi.org/10.1073/pnas.96.20.10976
Groll M, Huber R (2004) Inhibitors of the eukaryotic 20S proteasome core particle: a structural approach. Biochim Biophys Acta 1695(1–3):33–44. https://doi.org/10.1016/j.bbamcr.2004.09.025
Grune T, Jung T, Merker K, Davies KJ (2004) Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int J Biochem Cell Biol 36(12):2519–2530. https://doi.org/10.1016/j.biocel.2004.04.020
Gu ZC, Wu E, Sailer C, Jando J, Styles E, Eisenkolb I, Kuschel M, Bitschar K, Wang X, Huang L, Vissa A, Yip CM, Yedidi RS, Friesen H, Enenkel C (2017) Ubiquitin orchestrates proteasome dynamics between proliferation and quiescence in yeast. Mol Biol Cell 28(19):2479–2491. https://doi.org/10.1091/mbc.E17-03-0162
Guerrero C, Tagwerker C, Kaiser P, Huang L (2006) An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol Cell Proteomics 5(2):366–378. https://doi.org/10.1074/mcp.M500303-MCP200
Guo Q, Lehmer C, Martinez-Sanchez A, Rudack T, Beck F, Hartmann H, Perez-Berlanga M, Frottin F, Hipp MS, Hartl FU, Edbauer D, Baumeister W, Fernandez-Busnadiego R (2018) In situ structure of neuronal C9orf72 Poly-GA aggregates reveals proteasome recruitment. Cell 172(4):696–705 e612. https://doi.org/10.1016/j.cell.2017.12.030
Guo X, Engel JL, Xiao J, Tagliabracci VS, Wang X, Huang L, Dixon JE (2011) UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity. Proc Natl Acad Sci U S A 108(46):18649–18654. https://doi.org/10.1073/pnas.1113170108
Guterman A, Glickman MH (2004) Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem 279(3):1729–1738. https://doi.org/10.1074/jbc.M307050200
Haapasalo A, Viswanathan J, Kurkinen KM, Bertram L, Soininen H, Dantuma NP, Tanzi RE, Hiltunen M (2011) Involvement of ubiquilin-1 transcript variants in protein degradation and accumulation. Commun Integr Biol 4(4):428–432. https://doi.org/10.4161/cib.4.4.15283
Hahn JS, Neef DW, Thiele DJ (2006) A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol Microbiol 60(1):240–251. https://doi.org/10.1111/j.1365-2958.2006.05097.x
Hallermalm K, Seki K, Wei C, Castelli C, Rivoltini L, Kiessling R, Levitskaya J (2001) Tumor necrosis factor-alpha induces coordinated changes in major histocompatibility class I presentation pathway, resulting in increased stability of class I complexes at the cell surface. Blood 98(4):1108–1115. https://doi.org/10.1182/blood.v98.4.1108
Hamazaki J, Hirayama S, Murata S (2015) Redundant roles of Rpn10 and Rpn13 in recognition of ubiquitinated proteins and cellular homeostasis. PLoS Genet 11(7):e1005401. https://doi.org/10.1371/journal.pgen.1005401
Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J 25(19):4524–4536. https://doi.org/10.1038/sj.emboj.7601338
Han H, Fulcher JM, Dandey VP, Iwasa JH, Sundquist WI, Kay MS, Shen PS, Hill CP (2019) Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA + ATPases. Elife 8. https://doi.org/10.7554/elife.44071
Han H, Monroe N, Sundquist WI, Shen PS, Hill CP (2017) The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. Elife 6. https://doi.org/10.7554/elife.31324
Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127(1):99–111. https://doi.org/10.1016/j.cell.2006.07.038
Hanson PI, Whiteheart SW (2005) AAA + proteins: have engine, will work. Nat Rev Mol Cell Biol 6(7):519–529. https://doi.org/10.1038/nrm1684
Hanzelmann P, Schindelin H (2017) The interplay of cofactor interactions and post-translational modifications in the regulation of the AAA + ATPase p97. Front Mol Biosci 4:21. https://doi.org/10.3389/fmolb.2017.00021
Hanzelmann P, Stingele J, Hofmann K, Schindelin H, Raasi S (2010) The yeast E4 ubiquitin ligase Ufd2 interacts with the ubiquitin-like domains of Rad23 and Dsk2 via a novel and distinct ubiquitin-like binding domain. J Biol Chem 285(26):20390–20398. https://doi.org/10.1074/jbc.M110.112532
Haratake K, Sato A, Tsuruta F, Chiba T (2016) KIAA0368-deficiency affects disassembly of 26S proteasome under oxidative stress condition. J Biochem 159(6):609–618. https://doi.org/10.1093/jb/mvw006
Harding CV, France J, Song R, Farah JM, Chatterjee S, Iqbal M, Siman R (1995) Novel dipeptide aldehydes are proteasome inhibitors and block the MHC-I antigen-processing pathway. J Immunol 155(4):1767–1775
Harris JR (1968) Release of a macromolecular protein component from human erythrocyte ghosts. Biochim Biophys Acta 150(3):534–537. https://doi.org/10.1016/0005-2736(68)90157-0
Harshbarger W, Miller C, Diedrich C, Sacchettini J (2015) Crystal structure of the human 20S proteasome in complex with carfilzomib. Structure 23(2):418–424. https://doi.org/10.1016/j.str.2014.11.017
Haselbach D, Schrader J, Lambrecht F, Henneberg F, Chari A, Stark H (2017) Long-range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs. Nat Commun 8:15578. https://doi.org/10.1038/ncomms15578
Hayashi M, Ishibashi T, Tanaka K, Kasahara M (1997) The mouse genes encoding the third pair of beta-type proteasome subunits regulated reciprocally by IFN-gamma: structural comparison, chromosomal localization, and analysis of the promoter. J Immunol 159(6):2760–2770
He J, Kulkarni K, da Fonseca PC, Krutauz D, Glickman MH, Barford D, Morris EP (2012) The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric alpha-helical rings. Structure 20(3):513–521. https://doi.org/10.1016/j.str.2011.12.015
Hegde AN, Smith SG, Duke LM, Pourquoi A, Vaz S (2019) Perturbations of ubiquitin-proteasome-mediated proteolysis in aging and alzheimer’s disease. Front Aging Neurosci 11:324. https://doi.org/10.3389/fnagi.2019.00324
Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH (1997) The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J Biol Chem 272(40):25200–25209. https://doi.org/10.1074/jbc.272.40.25200
Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH (1993) PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem 268(7):5115–5120
Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH (1991) Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J 10(3):555–562
Heinen C, Acs K, Hoogstraten D, Dantuma NP (2011) C-terminal UBA domains protect ubiquitin receptors by preventing initiation of protein degradation. Nat Commun 2:191. https://doi.org/10.1038/ncomms1179
Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, Ashrafi K, Glickman MH, Rutter J (2010) A stress-responsive system for mitochondrial protein degradation. Mol Cell 40(3):465–480. https://doi.org/10.1016/j.molcel.2010.10.021
Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA (1980) Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci U S A 77(4):1783–1786. https://doi.org/10.1073/pnas.77.4.1783
Hershko A, Ciechanover A, Rose IA (1979) Resolution of the ATP-dependent proteolytic system from reticulocytes: a component that interacts with ATP. Proc Natl Acad Sci U S A 76(7):3107–3110. https://doi.org/10.1073/pnas.76.7.3107
Hilt W, Enenkel C, Gruhler A, Singer T, Wolf DH (1993) The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. Mutations link the proteasome to stress- and ubiquitin-dependent proteolysis. J Biol Chem 268(5):3479–3486
Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL (2005) Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121(7):1029–1041. https://doi.org/10.1016/j.cell.2005.04.012
Hirano H, Kimura Y, Kimura A (2016) Biological significance of co- and post-translational modifications of the yeast 26S proteasome. J Proteomics 134:37–46. https://doi.org/10.1016/j.jprot.2015.11.016
Hirano Y, Hayashi H, Iemura S, Hendil KB, Niwa S, Kishimoto T, Kasahara M, Natsume T, Tanaka K, Murata S (2006) Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol Cell 24(6):977–984. https://doi.org/10.1016/j.molcel.2006.11.015
Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, Natsume T, Tanaka K, Murata S (2005) A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 437(7063):1381–1385. https://doi.org/10.1038/nature04106
Hirano Y, Kaneko T, Okamoto K, Bai M, Yashiroda H, Furuyama K, Kato K, Tanaka K, Murata S (2008) Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J 27(16):2204–2213. https://doi.org/10.1038/emboj.2008.148
Hisamatsu H, Shimbara N, Saito Y, Kristensen P, Hendil KB, Fujiwara T, Takahashi E, Tanahashi N, Tamura T, Ichihara A, Tanaka K (1996) Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J Exp Med 183(4):1807–1816. https://doi.org/10.1084/jem.183.4.1807
Hishiya A, Iemura S, Natsume T, Takayama S, Ikeda K, Watanabe K (2006) A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J 25(3):554–564. https://doi.org/10.1038/sj.emboj.7600945
Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, Osborne G, Bates GP, Glickman MH, Trost M, Knebel A, Marchesi F, Kurz T (2016) UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell 166(4):935–949. https://doi.org/10.1016/j.cell.2016.07.001
Hnia K, Clausen T, Moog-Lutz C (2019) Shaping striated muscles with ubiquitin proteasome system in health and disease. Trends Mol Med. https://doi.org/10.1016/j.molmed.2019.05.008
Hoeller D, Dikic I (2009) Targeting the ubiquitin system in cancer therapy. Nature 458(7237):438–444. https://doi.org/10.1038/nature07960
Hoffman L, Rechsteiner M (1996) Nucleotidase activities of the 26 S proteasome and its regulatory complex. J Biol Chem 271(51):32538–32545
Hofmann K, Bucher P (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci 21(5):172–173
Hofmann K, Bucher P (1998) The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci 23(6):204–205. https://doi.org/10.1016/s0968-0004(98)01217-1
Hofmann RM, Pickart CM (2001) In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem 276(30):27936–27943. https://doi.org/10.1074/jbc.M103378200
Holzl H, Kapelari B, Kellermann J, Seemuller E, Sumegi M, Udvardy A, Medalia O, Sperling J, Muller SA, Engel A, Baumeister W (2000) The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol 150(1):119–130. https://doi.org/10.1083/jcb.150.1.119
Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S (2000) Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102(5):577–586. https://doi.org/10.1016/s0092-8674(00)00080-5
Hoppe T, Rape M, Jentsch S (2001) Membrane-bound transcription factors: regulated release by RIP or RUP. Curr Opin Cell Biol 13(3):344–348. https://doi.org/10.1016/s0955-0674(00)00218-0
Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N (2009) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20(7):1981–1991. https://doi.org/10.1091/mbc.E08-12-1248
Hospenthal MK, Freund SM, Komander D (2013) Assembly, analysis and architecture of atypical ubiquitin chains. Nat Struct Mol Biol 20(5):555–565. https://doi.org/10.1038/nsmb.2547
Hough R, Pratt G, Rechsteiner M (1986) Ubiquitin-lysozyme conjugates. Identification and characterization of an ATP-dependent protease from rabbit reticulocyte lysates. J Biol Chem 261(5):2400–2408
Hough R, Pratt G, Rechsteiner M (1987) Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem 262(17):8303–8313
Hoyt MA, Zich J, Takeuchi J, Zhang M, Govaerts C, Coffino P (2006) Glycine-alanine repeats impair proper substrate unfolding by the proteasome. EMBO J 25(8):1720–1729. https://doi.org/10.1038/sj.emboj.7601058
Hua Z, Vierstra RD (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62:299–334. https://doi.org/10.1146/annurev-arplant-042809-112256
Huang X, Luan B, Wu J, Shi Y (2016) An atomic structure of the human 26S proteasome. Nat Struct Mol Biol 23(9):778–785. https://doi.org/10.1038/nsmb.3273
Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, Groettrup M, Groll M (2012) Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 148(4):727–738. https://doi.org/10.1016/j.cell.2011.12.030
Huber EM, Groll M (2012) Inhibitors for the immuno- and constitutive proteasome: current and future trends in drug development. Angew Chem Int Ed Engl 51(35):8708–8720. https://doi.org/10.1002/anie.201201616
Huber EM, Heinemeyer W, Li X, Arendt CS, Hochstrasser M, Groll M (2016) A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome. Nat Commun 7:10900. https://doi.org/10.1038/ncomms10900
Hurley JH, Stenmark H (2011) Molecular mechanisms of ubiquitin-dependent membrane traffic. Annu Rev Biophys 40:119–142. https://doi.org/10.1146/annurev-biophys-042910-155404
Husnjak K, Dikic I (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem 81:291–322. https://doi.org/10.1146/annurev-biochem-051810-094654
Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453(7194):481–488. https://doi.org/10.1038/nature06926
Hussong SA, Kapphahn RJ, Phillips SL, Maldonado M, Ferrington DA (2010) Immunoproteasome deficiency alters retinal proteasome’s response to stress. J Neurochem 113(6):1481–1490. https://doi.org/10.1111/j.1471-4159.2010.06688.x
Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S (2004) Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem 279(37):38369–38378. https://doi.org/10.1074/jbc.M402468200
Inn KS, Gack MU, Tokunaga F, Shi M, Wong LY, Iwai K, Jung JU (2011) Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell 41(3):354–365. https://doi.org/10.1016/j.molcel.2010.12.029
Inobe T, Fishbain S, Prakash S, Matouschek A (2011) Defining the geometry of the two-component proteasome degron. Nat Chem Biol 7(3):161–167. https://doi.org/10.1038/nchembio.521
Iosefson O, Nager AR, Baker TA, Sauer RT (2015) Coordinated gripping of substrate by subunits of a AAA + proteolytic machine. Nat Chem Biol 11(3):201–206. https://doi.org/10.1038/nchembio.1732
Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, Kirkpatrick DS, Thomson TM, Finley D, Gygi SP, Crosas B (2010) Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol Cell 38(5):733–745. https://doi.org/10.1016/j.molcel.2010.05.001
Ishii T, Funakoshi M, Kobayashi H (2006) Yeast Pth2 is a UBL domain-binding protein that participates in the ubiquitin-proteasome pathway. EMBO J 25(23):5492–5503. https://doi.org/10.1038/sj.emboj.7601418
Isono E, Nishihara K, Saeki Y, Yashiroda H, Kamata N, Ge L, Ueda T, Kikuchi Y, Tanaka K, Nakano A, Toh-e A (2007) The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol Biol Cell 18(2):569–580. https://doi.org/10.1091/mbc.e06-07-0635
Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, Hegde RS (2016) Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol Cell 63(1):21–33. https://doi.org/10.1016/j.molcel.2016.05.020
Iwafune Y, Kawasaki H, Hirano H (2002) Electrophoretic analysis of phosphorylation of the yeast 20S proteasome. Electrophoresis 23(2):329–338. https://doi.org/10.1002/1522-2683(200202)23:2%3c329:AID-ELPS329%3e3.0.CO;2-1
Iwai K, Fujita H, Sasaki Y (2014) Linear ubiquitin chains: NF-kappaB signalling, cell death and beyond. Nat Rev Mol Cell Biol 15(8):503–508. https://doi.org/10.1038/nrm3836
Iwanczyk J, Sadre-Bazzaz K, Ferrell K, Kondrashkina E, Formosa T, Hill CP, Ortega J (2006) Structure of the Blm10–20 S proteasome complex by cryo-electron microscopy. Insights into the mechanism of activation of mature yeast proteasomes. J Mol Biol 363(3):648–659. https://doi.org/10.1016/j.jmb.2006.08.010
Iyer LM, Leipe DD, Koonin EV, Aravind L (2004) Evolutionary history and higher order classification of AAA + ATPases. J Struct Biol 146(1–2):11–31. https://doi.org/10.1016/j.jsb.2003.10.010
Jacobson AD, MacFadden A, Wu Z, Peng J, Liu CW (2014) Autoregulation of the 26S proteasome by in situ ubiquitination. Mol Biol Cell 25(12):1824–1835. https://doi.org/10.1091/mbc.E13-10-0585
Jagannath S, Barlogie B, Berenson J, Siegel D, Irwin D, Richardson PG, Niesvizky R, Alexanian R, Limentani SA, Alsina M, Adams J, Kauffman M, Esseltine DL, Schenkein DP, Anderson KC (2004) A phase 2 study of two doses of bortezomib in relapsed or refractory myeloma. Br J Haematol 127(2):165–172. https://doi.org/10.1111/j.1365-2141.2004.05188.x
James AB, Conway AM, Morris BJ (2006) Regulation of the neuronal proteasome by Zif268 (Egr1). J Neurosci 26(5):1624–1634. https://doi.org/10.1523/JNEUROSCI.4199-05.2006
Janse DM, Crosas B, Finley D, Church GM (2004) Localization to the proteasome is sufficient for degradation. J Biol Chem 279(20):21415–21420. https://doi.org/10.1074/jbc.M402954200
Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T (2002) Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol 4(2):134–139. https://doi.org/10.1038/ncb746
Jin L, Williamson A, Banerjee S, Philipp I, Rape M (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133(4):653–665. https://doi.org/10.1016/j.cell.2008.04.012
Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC (2009) Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol 187(6):875–888. https://doi.org/10.1083/jcb.200908115
Kaganovich D, Kopito R, Frydman J (2008) Misfolded proteins partition between two distinct quality control compartments. Nature 454(7208):1088–1095. https://doi.org/10.1038/nature07195
Kaiser SE, Riley BE, Shaler TA, Trevino RS, Becker CH, Schulman H, Kopito RR (2011) Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat Methods 8(8):691–696. https://doi.org/10.1038/nmeth.1649
Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S (2009) Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell 137(5):914–925. https://doi.org/10.1016/j.cell.2009.05.008
Kaplun L, Tzirkin R, Bakhrat A, Shabek N, Ivantsiv Y, Raveh D (2005) The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol Cell Biol 25(13):5355–5362. https://doi.org/10.1128/MCB.25.13.5355-5362.2005
Karmon O, Ben Aroya S (2019) Spatial organization of proteasome aggregates in the regulation of proteasome homeostasis. Front Mol Biosci 6:150. https://doi.org/10.3389/fmolb.2019.00150
Kay LE (2011) Solution NMR spectroscopy of supra-molecular systems, why bother?. A methyl-TROSY view. J Magn Reson 210(2):159–170. https://doi.org/10.1016/j.jmr.2011.03.008
Keller JN, Hanni KB, Markesbery WR (2000) Impaired proteasome function in Alzheimer’s disease. J Neurochem 75(1):436–439. https://doi.org/10.1046/j.1471-4159.2000.0750436.x
Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, Jovanovic M, Ellis SR, Fulco CP, Engreitz JM, Schutz S, Lian J, Gripp KW, Weinberg OK, Pinkus GS, Gehrke L, Regev A, Lander ES, Gazda HT, Lee WY, Panse VG, Carr SA, Sankaran VG (2018) Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173(1):90–103 e119. https://doi.org/10.1016/j.cell.2018.02.036
Khare S, Nagle AS, Biggart A, Lai YH, Liang F, Davis LC, Barnes SW, Mathison CJ, Myburgh E, Gao MY, Gillespie JR, Liu X, Tan JL, Stinson M, Rivera IC, Ballard J, Yeh V, Groessl T, Federe G, Koh HX, Venable JD, Bursulaya B, Shapiro M, Mishra PK, Spraggon G, Brock A, Mottram JC, Buckner FS, Rao SP, Wen BG, Walker JR, Tuntland T, Molteni V, Glynne RJ, Supek F (2016) Proteasome inhibition for treatment of leishmaniasis, Chagas disease and sleeping sickness. Nature 537(7619):229–233. https://doi.org/10.1038/nature19339
Khmelinskii A, Blaszczak E, Pantazopoulou M, Fischer B, Omnus DJ, Le Dez G, Brossard A, Gunnarsson A, Barry JD, Meurer M, Kirrmaier D, Boone C, Huber W, Rabut G, Ljungdahl PO, Knop M (2014) Protein quality control at the inner nuclear membrane. Nature 516(7531):410–413. https://doi.org/10.1038/nature14096
Kikuchi J, Iwafune Y, Akiyama T, Okayama A, Nakamura H, Arakawa N, Kimura Y, Hirano H (2010) Co- and post-translational modifications of the 26S proteasome in yeast. Proteomics 10(15):2769–2779. https://doi.org/10.1002/pmic.200900283
Kim DY, Scalf M, Smith LM, Vierstra RD (2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25(5):1523–1540. https://doi.org/10.1105/tpc.112.108613
Kim HT, Goldberg AL (2017) The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis. J Biol Chem 292(23):9830–9839. https://doi.org/10.1074/jbc.M116.763128
Kim HT, Goldberg AL (2018) UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells. Proc Natl Acad Sci U S A 115(50):E11642–E11650. https://doi.org/10.1073/pnas.1808731115
Kim I, Ahn J, Liu C, Tanabe K, Apodaca J, Suzuki T, Rao H (2006) The Png1-Rad23 complex regulates glycoprotein turnover. J Cell Biol 172(2):211–219. https://doi.org/10.1083/jcb.200507149
Kim I, Mi K, Rao H (2004) Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell 15(7):3357–3365. https://doi.org/10.1091/mbc.e03-11-0835
Kim S, Saeki Y, Fukunaga K, Suzuki A, Takagi K, Yamane T, Tanaka K, Mizushima T, Kato K (2010) Crystal structure of yeast rpn14, a chaperone of the 19 S regulatory particle of the proteasome. J Biol Chem 285(20):15159–15166. https://doi.org/10.1074/jbc.M110.104042
Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44(2):325–340. https://doi.org/10.1016/j.molcel.2011.08.025
Kim YC, DeMartino GN (2011) C termini of proteasomal ATPases play nonequivalent roles in cellular assembly of mammalian 26 S proteasome. J Biol Chem 286(30):26652–26666. https://doi.org/10.1074/jbc.M111.246793
Kim YC, Snoberger A, Schupp J, Smith DM (2015) ATP binding to neighbouring subunits and intersubunit allosteric coupling underlie proteasomal ATPase function. Nat Commun 6:8520. https://doi.org/10.1038/ncomms9520
Kimonis VE, Fulchiero E, Vesa J, Watts G (2008) VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys Acta 1782(12):744–748. https://doi.org/10.1016/j.bbadis.2008.09.003
Kimura A, Kato Y, Hirano H (2012) N-myristoylation of the Rpt2 subunit regulates intracellular localization of the yeast 26S proteasome. Biochemistry 51(44):8856–8866. https://doi.org/10.1021/bi3007862
Kimura A, Kurata Y, Nakabayashi J, Kagawa H, Hirano H (2016) N-Myristoylation of the Rpt2 subunit of the yeast 26S proteasome is implicated in the subcellular compartment-specific protein quality control system. J Proteomics 130:33–41. https://doi.org/10.1016/j.jprot.2015.08.021
Kimura Y, Kurata Y, Ishikawa A, Okayama A, Kamita M, Hirano H (2013) N-Terminal methylation of proteasome subunit Rpt1 in yeast. Proteomics 13(21):3167–3174. https://doi.org/10.1002/pmic.201300207
King RW, Deshaies RJ, Peters JM, Kirschner MW (1996) How proteolysis drives the cell cycle. Science 274(5293):1652–1659. https://doi.org/10.1126/science.274.5293.1652
Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25(20):4877–4887. https://doi.org/10.1038/sj.emboj.7601360
Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol 8(7):700–710. https://doi.org/10.1038/ncb1436
Kisselev AF (2013) A novel bullet hits the proteasome. Cancer Cell 24(6):691–693. https://doi.org/10.1016/j.ccr.2013.11.016
Kisselev AF, Akopian TN, Castillo V, Goldberg AL (1999) Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol Cell 4(3):395–402. https://doi.org/10.1016/s1097-2765(00)80341-x
Kisselev AF, Akopian TN, Goldberg AL (1998) Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J Biol Chem 273(4):1982–1989. https://doi.org/10.1074/jbc.273.4.1982
Kisselev AF, Goldberg AL (2001) Proteasome inhibitors: from research tools to drug candidates. Chem Biol 8(8):739–758. https://doi.org/10.1016/s1074-5521(01)00056-4
Kleijnen MF, Roelofs J, Park S, Hathaway NA, Glickman M, King RW, Finley D (2007) Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat Struct Mol Biol 14(12):1180–1188. https://doi.org/10.1038/nsmb1335
Klionsky DJ, Schulman BA (2014) Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol 21(4):336–345. https://doi.org/10.1038/nsmb.2787
Kloetzel PM (2001) Antigen processing by the proteasome. Nat Rev Mol Cell Biol 2(3):179–187. https://doi.org/10.1038/35056572
Knowlton JR, Johnston SC, Whitby FG, Realini C, Zhang Z, Rechsteiner M, Hill CP (1997) Structure of the proteasome activator REGalpha (PA28alpha). Nature 390(6660):639–643. https://doi.org/10.1038/37670
Ko HS, Uehara T, Tsuruma K, Nomura Y (2004) Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett 566(1–3):110–114. https://doi.org/10.1016/j.febslet.2004.04.031
Kock M, Nunes MM, Hemann M, Kube S, Dohmen RJ, Herzog F, Ramos PC, Wendler P (2015) Proteasome assembly from 15S precursors involves major conformational changes and recycling of the Pba1-Pba2 chaperone. Nat Commun 6:6123. https://doi.org/10.1038/ncomms7123
Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96(5):635–644. https://doi.org/10.1016/s0092-8674(00)80574-7
Komander D, Clague MJ, Urbe S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10(8):550–563. https://doi.org/10.1038/nrm2731
Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81:203–229. https://doi.org/10.1146/annurev-biochem-060310-170328
Kopp F, Dahlmann B, Kuehn L (2001) Reconstitution of hybrid proteasomes from purified PA700-20 S complexes and PA28alphabeta activator: ultrastructure and peptidase activities. J Mol Biol 313(3):465–471. https://doi.org/10.1006/jmbi.2001.5063
Kors S, Geijtenbeek K, Reits E, Schipper-Krom S (2019) Regulation of proteasome activity by (Post-)transcriptional mechanisms. Front Mol Biosci 6:48. https://doi.org/10.3389/fmolb.2019.00048
Kraut DA, Israeli E, Schrader EK, Patil A, Nakai K, Nanavati D, Inobe T, Matouschek A (2012) Sequence- and species-dependence of proteasomal processivity. ACS Chem Biol 7(8):1444–1453. https://doi.org/10.1021/cb3001155
Kraut DA, Matouschek A (2011) Proteasomal degradation from internal sites favors partial proteolysis via remote domain stabilization. ACS Chem Biol 6(10):1087–1095. https://doi.org/10.1021/cb2002285
Kravtsova-Ivantsiv Y, Ciechanover A (2012) Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci 125(Pt 3):539–548. https://doi.org/10.1242/jcs.093567
Kravtsova-Ivantsiv Y, Cohen S, Ciechanover A (2009) Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Mol Cell 33(4):496–504. https://doi.org/10.1016/j.molcel.2009.01.023
Kristariyanto YA, Abdul Rehman SA, Campbell DG, Morrice NA, Johnson C, Toth R, Kulathu Y (2015a) K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin. Mol Cell 58(1):83–94. https://doi.org/10.1016/j.molcel.2015.01.041
Kristariyanto YA, Choi SY, Rehman SA, Ritorto MS, Campbell DG, Morrice NA, Toth R, Kulathu Y (2015b) Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations. Biochem J 467(2):345–352. https://doi.org/10.1042/BJ20141502
Kruegel U, Robison B, Dange T, Kahlert G, Delaney JR, Kotireddy S, Tsuchiya M, Tsuchiyama S, Murakami CJ, Schleit J, Sutphin G, Carr D, Tar K, Dittmar G, Kaeberlein M, Kennedy BK, Schmidt M (2011) Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet 7(9):e1002253. https://doi.org/10.1371/journal.pgen.1002253
Krzywda S, Brzozowski AM, Higashitsuji H, Fujita J, Welchman R, Dawson S, Mayer RJ, Wilkinson AJ (2004) The crystal structure of gankyrin, an oncoprotein found in complexes with cyclin-dependent kinase 4, a 19 S proteasomal ATPase regulator, and the tumor suppressors Rb and p53. J Biol Chem 279(2):1541–1545. https://doi.org/10.1074/jbc.M310265200
Kuhnle S, Martinez-Noel G, Leclere F, Hayes SD, Harper JW, Howley PM (2018) Angelman syndrome-associated point mutations in the Zn(2 +)-binding N-terminal (AZUL) domain of UBE3A ubiquitin ligase inhibit binding to the proteasome. J Biol Chem 293(47):18387–18399. https://doi.org/10.1074/jbc.RA118.004653
Kulathu Y, Komander D (2012) Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol 13(8):508–523. https://doi.org/10.1038/nrm3394
Kuo CL, Goldberg AL (2017) Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c. Proc Natl Acad Sci U S A 114(17):E3404–E3413. https://doi.org/10.1073/pnas.1701734114
Kusmierczyk AR, Hochstrasser M (2008) Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol Chem 389(9):1143–1151. https://doi.org/10.1515/BC.2008.130
Kwon YD, Nagy I, Adams PD, Baumeister W, Jap BK (2004) Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly. J Mol Biol 335(1):233–245. https://doi.org/10.1016/j.jmb.2003.08.029
Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM (2002) A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416(6882):763–767. https://doi.org/10.1038/416763a
Lam YA, Xu W, DeMartino GN, Cohen RE (1997) Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385(6618):737–740. https://doi.org/10.1038/385737a0
Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482(7384):186–191. https://doi.org/10.1038/nature10774
Laporte D, Salin B, Daignan-Fornier B, Sagot I (2008) Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol 181(5):737–745. https://doi.org/10.1083/jcb.200711154
Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A 109(5):1380–1387. https://doi.org/10.1073/pnas.1120559109
Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A (2007) 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell 27(4):660–674. https://doi.org/10.1016/j.molcel.2007.06.025
Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467(7312):179–184. https://doi.org/10.1038/nature09299
Lee BH, Lu Y, Prado MA, Shi Y, Tian G, Sun S, Elsasser S, Gygi SP, King RW, Finley D (2016) USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 532(7599):398–401. https://doi.org/10.1038/nature17433
Lee CK, Klopp RG, Weindruch R, Prolla TA (1999) Gene expression profile of aging and its retardation by caloric restriction. Science 285(5432):1390–1393. https://doi.org/10.1126/science.285.5432.1390
Lee D, Takayama S, Goldberg AL (2018) ZFAND5/ZNF216 is an activator of the 26S proteasome that stimulates overall protein degradation. Proc Natl Acad Sci U S A 115(41):E9550–E9559. https://doi.org/10.1073/pnas.1809934115
Lee DH, Goldberg AL (1998) Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 8(10):397–403. https://doi.org/10.1016/s0962-8924(98)01346-4
Lee SY, De la Mota-Peynado A, Roelofs J (2011) Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. J Biol Chem 286(42):36641–36651. https://doi.org/10.1074/jbc.M111.280875
Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D (2002) Multiple associated proteins regulate proteasome structure and function. Mol Cell 10(3):495–507. https://doi.org/10.1016/s1097-2765(02)00638-x
Lehmann A, Janek K, Braun B, Kloetzel PM, Enenkel C (2002) 20 S proteasomes are imported as precursor complexes into the nucleus of yeast. J Mol Biol 317(3):401–413. https://doi.org/10.1006/jmbi.2002.5443
Lehmann A, Niewienda A, Jechow K, Janek K, Enenkel C (2010) Ecm29 fulfils quality control functions in proteasome assembly. Mol Cell 38(6):879–888. https://doi.org/10.1016/j.molcel.2010.06.016
Leone P, Shin EC, Perosa F, Vacca A, Dammacco F, Racanelli V (2013) MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J Natl Cancer Inst 105(16):1172–1187. https://doi.org/10.1093/jnci/djt184
Levine B, Kroemer G (2019) Biological functions of autophagy genes: a disease perspective. Cell 176(1–2):11–42. https://doi.org/10.1016/j.cell.2018.09.048
Leznicki P, Kulathu Y (2017) Mechanisms of regulation and diversification of deubiquitylating enzyme function. J Cell Sci 130(12):1997–2006. https://doi.org/10.1242/jcs.201855
Li F, Tian G, Langager D, Sokolova V, Finley D, Park S (2017) Nucleotide-dependent switch in proteasome assembly mediated by the Nas6 chaperone. Proc Natl Acad Sci U S A 114(7):1548–1553. https://doi.org/10.1073/pnas.1612922114
Li H, O’Donoghue AJ, van der Linden WA, Xie SC, Yoo E, Foe IT, Tilley L, Craik CS, da Fonseca PC, Bogyo M (2016a) Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature 530(7589):233–236. https://doi.org/10.1038/nature16936
Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X (2011) Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest 121(9):3689–3700. https://doi.org/10.1172/JCI45709
Li X, Amazit L, Long W, Lonard DM, Monaco JJ, O’Malley BW (2007a) Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell 26(6):831–842. https://doi.org/10.1016/j.molcel.2007.05.028
Li X, Demartino GN (2009) Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem J 421(3):397–404. https://doi.org/10.1042/BJ20090528
Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M (2007b) beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J 26(9):2339–2349. https://doi.org/10.1038/sj.emboj.7601681
Li X, Li Y, Arendt CS, Hochstrasser M (2016b) Distinct elements in the proteasomal beta5 subunit propeptide required for autocatalytic processing and proteasome assembly. J Biol Chem 291(4):1991–2003. https://doi.org/10.1074/jbc.M115.677047
Li X, Thompson D, Kumar B, DeMartino GN (2014) Molecular and cellular roles of PI31 (PSMF1) protein in regulation of proteasome function. J Biol Chem 289(25):17392–17405. https://doi.org/10.1074/jbc.M114.561183
Li X, Wang CE, Huang S, Xu X, Li XJ, Li H, Li S (2010) Inhibiting the ubiquitin-proteasome system leads to preferential accumulation of toxic N-terminal mutant huntingtin fragments. Hum Mol Genet 19(12):2445–2455. https://doi.org/10.1093/hmg/ddq127
Li YF, Wang X (2011) The role of the proteasome in heart disease. Biochim Biophys Acta 1809(2):141–149. https://doi.org/10.1016/j.bbagrm.2010.09.001
Liang RY, Chen L, Ko BT, Shen YH, Li YT, Chen BR, Lin KT, Madura K, Chuang SM (2014) Rad23 interaction with the proteasome is regulated by phosphorylation of its ubiquitin-like (UbL) domain. J Mol Biol 426(24):4049–4060. https://doi.org/10.1016/j.jmb.2014.10.004
Lin G, Li D, de Carvalho LP, Deng H, Tao H, Vogt G, Wu K, Schneider J, Chidawanyika T, Warren JD, Li H, Nathan C (2009) Inhibitors selective for mycobacterial versus human proteasomes. Nature 461(7264):621–626. https://doi.org/10.1038/nature08357
Lin G, Mao D, Bellen HJ (2017) Amyotrophic lateral sclerosis pathogenesis converges on defects in protein homeostasis associated with TDP-43 mislocalization and proteasome-mediated degradation overload. Curr Top Dev Biol 121:111–171. https://doi.org/10.1016/bs.ctdb.2016.07.004
Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, Renatus M, Fischer ES, Thoma NH (2014) Crystal structure of the human COP9 signalosome. Nature 512(7513):161–165. https://doi.org/10.1038/nature13566
Lipinszki Z, Kovacs L, Deak P, Udvardy A (2012) Ubiquitylation of drosophila p54/Rpn10/S5a regulates its interaction with the UBA-UBL polyubiquitin receptors. Biochemistry 51(12):2461–2470. https://doi.org/10.1021/bi3001006
Liu X, Xiao W, Zhang Y, Wiley SE, Zuo T, Zheng Y, Chen N, Chen L, Wang X, Zheng Y, Huang L, Lin S, Murphy AN, Dixon JE, Xu P, Guo X (2020) Reversible phosphorylation of Rpn1 regulates 26S proteasome assembly and function. Proc Natl Acad Sci U S A 117(1):328–336. https://doi.org/10.1073/pnas.1912531117
Liu Y, Lu L, Hettinger CL, Dong G, Zhang D, Rezvani K, Wang X, Wang H (2014) Ubiquilin-1 protects cells from oxidative stress and ischemic stroke caused tissue injury in mice. J Neurosci 34(8):2813–2821. https://doi.org/10.1523/JNEUROSCI.3541-13.2014
Liu Z, Dong X, Yi HW, Yang J, Gong Z, Wang Y, Liu K, Zhang WP, Tang C (2019) Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13. Cell Discov 5:19. https://doi.org/10.1038/s41421-019-0089-7
Liu Z, Gong Z, Jiang WX, Yang J, Zhu WK, Guo DC, Zhang WP, Liu ML, Tang C (2015) Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition. Elife 4. https://doi.org/10.7554/elife.05767
Livneh I, Cohen-Kaplan V, Cohen-Rosenzweig C, Avni N, Ciechanover A (2016) The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. Cell Res 26(8):869–885. https://doi.org/10.1038/cr.2016.86
Lokireddy S, Kukushkin NV, Goldberg AL (2015) cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc Natl Acad Sci U S A 112(52):E7176–E7185. https://doi.org/10.1073/pnas.1522332112
Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 268(5210):533–539. https://doi.org/10.1126/science.7725097
Lu H, Zong C, Wang Y, Young GW, Deng N, Souda P, Li X, Whitelegge J, Drews O, Yang PY, Ping P (2008) Revealing the dynamics of the 20 S proteasome phosphoproteome: a combined CID and electron transfer dissociation approach. Mol Cell Proteomics 7(11):2073–2089. https://doi.org/10.1074/mcp.M800064-MCP200
Lu K, Psakhye I, Jentsch S (2014) Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 158(3):549–563. https://doi.org/10.1016/j.cell.2014.05.048
Lu X, Ebelle DL, Matsuo H, Walters KJ (2020) An extended conformation for K48 ubiquitin chains revealed by the hRpn2:Rpn13:K48-Diubiquitin structure. Structure. https://doi.org/10.1016/j.str.2020.02.007
Lu X, Nowicka U, Sridharan V, Liu F, Randles L, Hymel D, Dyba M, Tarasov SG, Tarasova NI, Zhao XZ, Hamazaki J, Murata S, Burke TR Jr, Walters KJ (2017a) Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets. Nat Commun 8:15540. https://doi.org/10.1038/ncomms15540
Lu Y, Lee BH, King RW, Finley D, Kirschner MW (2015) Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science 348(6231):1250834. https://doi.org/10.1126/science.1250834
Lu Y, Wu J, Dong Y, Chen S, Sun S, Ma YB, Ouyang Q, Finley D, Kirschner MW, Mao Y (2017b) Conformational landscape of the p 28-Bound human proteasome regulatory particle. Mol Cell 67(2):322–333 e326. https://doi.org/10.1016/j.molcel.2017.06.007
Luan B, Huang X, Wu J, Mei Z, Wang Y, Xue X, Yan C, Wang J, Finley DJ, Shi Y, Wang F (2016) Structure of an endogenous yeast 26S proteasome reveals two major conformational states. Proc Natl Acad Sci U S A 113(10):2642–2647. https://doi.org/10.1073/pnas.1601561113
Lupas A, Zwickl P, Wenzel T, Seemuller E, Baumeister W (1995) Structure and function of the 20S proteasome and of its regulatory complexes. Cold Spring Harb Symp Quant Biol 60:515–524. https://doi.org/10.1101/sqb.1995.060.01.055
Lupas AN, Martin J (2002) AAA proteins. Curr Opin Struct Biol 12(6):746–753
Ma CP, Slaughter CA, DeMartino GN (1992) Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J Biol Chem 267(15):10515–10523
Ma M, Liu ZL (2010) Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genom 11:660. https://doi.org/10.1186/1471-2164-11-660
Ma W, Lehner PJ, Cresswell P, Pober JS, Johnson DR (1997) Interferon-gamma rapidly increases peptide transporter (TAP) subunit expression and peptide transport capacity in endothelial cells. J Biol Chem 272(26):16585–16590. https://doi.org/10.1074/jbc.272.26.16585
Machiels BM, Henfling ME, Gerards WL, Broers JL, Bloemendal H, Ramaekers FC, Schutte B (1997) Detailed analysis of cell cycle kinetics upon proteasome inhibition. Cytometry 28(3):243–252
Magnaghi P, D’Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, Polucci P, Ballinari D, Perrera C, Leone A, Cervi G, Casale E, Xiao Y, Wong C, Anderson DJ, Galvani A, Donati D, O’Brien T, Jackson PK, Isacchi A (2013) Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol 9(9):548–556. https://doi.org/10.1038/nchembio.1313
Majumder P, Rudack T, Beck F, Danev R, Pfeifer G, Nagy I, Baumeister W (2019) Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle. Proc Natl Acad Sci U S A 116(2):534–539. https://doi.org/10.1073/pnas.1817752116
Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S (2012) Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell 23(16):3041–3056. https://doi.org/10.1091/mbc.E12-03-0194
Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H (1999) Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett 450(1–2):27–34. https://doi.org/10.1016/s0014-5793(99)00467-6
Mansour W, Nakasone MA, von Delbruck M, Yu Z, Krutauz D, Reis N, Kleifeld O, Sommer T, Fushman D, Glickman MH (2015) Disassembly of Lys11 and mixed linkage polyubiquitin conjugates provides insights into function of proteasomal deubiquitinases Rpn11 and Ubp6. J Biol Chem 290(8):4688–4704. https://doi.org/10.1074/jbc.M114.568295
Maqbool A, Hughes RK, Dagdas YF, Tregidgo N, Zess E, Belhaj K, Round A, Bozkurt TO, Kamoun S, Banfield MJ (2016) Structural basis of host autophagy-related protein 8 (ATG8) binding by the irish potato famine pathogen effector protein PexRD54. J Biol Chem 291(38):20270–20282. https://doi.org/10.1074/jbc.M116.744995
Marshall RS, Hua Z, Mali S, McLoughlin F, Vierstra RD (2019) ATG8-Binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 177(3):766–781 e724. https://doi.org/10.1016/j.cell.2019.02.009
Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD (2015) Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/Ubiquitin receptor RPN10 in arabidopsis. Mol Cell 58(6):1053–1066. https://doi.org/10.1016/j.molcel.2015.04.023
Marshall RS, McLoughlin F, Vierstra RD (2016) Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Rep 16(6):1717–1732. https://doi.org/10.1016/j.celrep.2016.07.015
Marshall RS, Vierstra RD (2015) Eat or be eaten: the autophagic plight of inactive 26S proteasomes. Autophagy 11(10):1927–1928. https://doi.org/10.1080/15548627.2015.1078961
Marshall RS, Vierstra RD (2018a) Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol 69:173–208. https://doi.org/10.1146/annurev-arplant-042817-040606
Marshall RS, Vierstra RD (2018b) Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. Elife 7. https://doi.org/10.7554/elife.34532
Marshall RS, Vierstra RD (2019) Dynamic regulation of the 26S proteasome: from synthesis to degradation. Front Mol Biosci 6:40. https://doi.org/10.3389/fmolb.2019.00040
Martin A, Baker TA, Sauer RT (2008) Pore loops of the AAA + ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol 15(11):1147–1151. https://doi.org/10.1038/nsmb.1503
Martinez-Fonts K, Matouschek A (2016) A rapid and versatile method for generating proteins with defined ubiquitin chains. Biochemistry 55(12):1898–1908. https://doi.org/10.1021/acs.biochem.5b01310
Martinez-Noel G, Galligan JT, Sowa ME, Arndt V, Overton TM, Harper JW, Howley PM (2012) Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol Cell Biol 32(15):3095–3106. https://doi.org/10.1128/MCB.00201-12
Martinez-Fonts K, Davis C, Tomita T, Elsasser S, Nager AR, Shi Y, Finley D, Matouschek A (2020) The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat Commun 11(1):477. https://doi.org/10.1038/s41467-019-13906-8
Matiuhin Y, Kirkpatrick DS, Ziv I, Kim W, Dakshinamurthy A, Kleifeld O, Gygi SP, Reis N, Glickman MH (2008) Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell 32(3):415–425. https://doi.org/10.1016/j.molcel.2008.10.011
Matyskiela ME, Lander GC, Martin A (2013) Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol 20(7):781–788. https://doi.org/10.1038/nsmb.2616
Mayer RJ (2000) The meteoric rise of regulated intracellular proteolysis. Nat Rev Mol Cell Biol 1(2):145–148. https://doi.org/10.1038/35040090
Mayr J, Seemuller E, Muller SA, Engel A, Baumeister W (1998) Late events in the assembly of 20S proteasomes. J Struct Biol 124(2–3):179–188. https://doi.org/10.1006/jsbi.1998.4068
Maytal-Kivity V, Reis N, Hofmann K, Glickman MH (2002) MPN + , a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem 3:28. https://doi.org/10.1186/1471-2091-3-28
Mc Cormack T, Baumeister W, Grenier L, Moomaw C, Plamondon L, Pramanik B, Slaughter C, Soucy F, Stein R, Zuhl F, Dick L (1997) Active site-directed inhibitors of Rhodococcus 20 S proteasome. Kinetics and mechanism. J Biol Chem 272(42):26103–26109. https://doi.org/10.1074/jbc.272.42.26103
McCutchen-Maloney SL, Matsuda K, Shimbara N, Binns DD, Tanaka K, Slaughter CA, DeMartino GN (2000) cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J Biol Chem 275(24):18557–18565. https://doi.org/10.1074/jbc.M001697200
McNaught KS, Mytilineou C, Jnobaptiste R, Yabut J, Shashidharan P, Jennert P, Olanow CW (2002) Impairment of the ubiquitin-proteasome system causes dopaminergic cell death and inclusion body formation in ventral mesencephalic cultures. J Neurochem 81(2):301–306. https://doi.org/10.1046/j.1471-4159.2002.00821.x
McNaught KS, Perl DP, Brownell AL, Olanow CW (2004) Systemic exposure to proteasome inhibitors causes a progressive model of Parkinson’s disease. Ann Neurol 56(1):149–162. https://doi.org/10.1002/ana.20186
Meyer H, Bug M, Bremer S (2012) Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14(2):117–123. https://doi.org/10.1038/ncb2407
Meyer HJ, Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157(4):910–921. https://doi.org/10.1016/j.cell.2014.03.037
Meyer-Schwesinger C (2019) The ubiquitin-proteasome system in kidney physiology and disease. Nat Rev Nephrol 15(7):393–411. https://doi.org/10.1038/s41581-019-0148-1
Miller SB, Mogk A, Bukau B (2015) Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J Mol Biol 427(7):1564–1574. https://doi.org/10.1016/j.jmb.2015.02.006
Minis A, Rodriguez JA, Levin A, Liu K, Govek EE, Hatten ME, Steller H (2019) The proteasome regulator PI31 is required for protein homeostasis, synapse maintenance, and neuronal survival in mice. Proc Natl Acad Sci U S A 116 (49):24639–24650. https://doi.org/10.1073/pnas.1911921116
Mishto M, Bellavista E, Santoro A, Stolzing A, Ligorio C, Nacmias B, Spazzafumo L, Chiappelli M, Licastro F, Sorbi S, Pession A, Ohm T, Grune T, Franceschi C (2006) Immunoproteasome and LMP2 polymorphism in aged and Alzheimer’s disease brains. Neurobiol Aging 27(1):54–66. https://doi.org/10.1016/j.neurobiolaging.2004.12.004
Monroe N, Han H, Shen PS, Sundquist WI, Hill CP (2017) Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase. Elife 6. https://doi.org/10.7554/elife.24487
Moran E, Carbone F, Augusti V, Patrone F, Ballestrero A, Nencioni A (2012) Proteasome inhibitors as immunosuppressants: biological rationale and clinical experience. Semin Hematol 49(3):270–276. https://doi.org/10.1053/j.seminhematol.2012.04.004
Morris GP, Clark IA, Vissel B (2018) Questions concerning the role of amyloid-beta in the definition, aetiology and diagnosis of Alzheimer’s disease. Acta Neuropathol 136(5):663–689. https://doi.org/10.1007/s00401-018-1918-8
Motosugi R, Murata S (2019) Dynamic regulation of proteasome expression. Front Mol Biosci 6:30. https://doi.org/10.3389/fmolb.2019.00030
Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, Sylvain C, Ring ER, Shields J, Jiang J, Shwonek P, Parlati F, Demo SD, Bennett MK, Kirk CJ, Groettrup M (2009) A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med 15(7):781–787. https://doi.org/10.1038/nm.1978
Mullane K, Williams M (2019) Preclinical models of alzheimer’s disease: relevance and translational validity. Curr Protoc Pharmacol 84(1):e57. https://doi.org/10.1002/cpph.57
Muller AU, Weber-Ban E (2019) The bacterial proteasome at the core of diverse degradation pathways. Front Mol Biosci 6:23. https://doi.org/10.3389/fmolb.2019.00023
Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A (1992) Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360(6404):597–599. https://doi.org/10.1038/360597a0
Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K (2007) Regulation of CD8 + T cell development by thymus-specific proteasomes. Science 316(5829):1349–1353. https://doi.org/10.1126/science.1141915
Murata S, Takahama Y, Kasahara M, Tanaka K (2018) The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol 19(9):923–931. https://doi.org/10.1038/s41590-018-0186-z
Murata S, Yashiroda H, Tanaka K (2009) Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol 10(2):104–115. https://doi.org/10.1038/nrm2630
Nahar A, Fu X, Polovin G, Orth JD, Park S (2019) Two alternative mechanisms regulate the onset of chaperone-mediated assembly of the proteasomal ATPases. J Biol Chem 294(16):6562–6577. https://doi.org/10.1074/jbc.RA118.006298
Nakamura M, Tokunaga F, Sakata S, Iwai K (2006) Mutual regulation of conventional protein kinase C and a ubiquitin ligase complex. Biochem Biophys Res Commun 351(2):340–347. https://doi.org/10.1016/j.bbrc.2006.09.163
Nakamura Y, Nakano K, Umehara T, Kimura M, Hayashizaki Y, Tanaka A, Horikoshi M, Padmanabhan B, Yokoyama S (2007a) Structure of the oncoprotein gankyrin in complex with S6 ATPase of the 26S proteasome. Structure 15(2):179–189. https://doi.org/10.1016/j.str.2006.11.015
Nakamura Y, Umehara T, Tanaka A, Horikoshi M, Padmanabhan B, Yokoyama S (2007b) Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem Biophys Res Commun 359(3):503–509. https://doi.org/10.1016/j.bbrc.2007.05.138
Nederlof PM, Wang HR, Baumeister W (1995) Nuclear localization signals of human and Thermoplasma proteasomal alpha subunits are functional in vitro. Proc Natl Acad Sci U S A 92(26):12060–12064. https://doi.org/10.1073/pnas.92.26.12060
Nemec AA, Howell LA, Peterson AK, Murray MA, Tomko RJ Jr (2017) Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. J Biol Chem 292(52):21466–21480. https://doi.org/10.1074/jbc.M117.817999
Nemec AA, Peterson AK, Warnock JL, Reed RG, Tomko RJ, Jr. (2019) an allosteric interaction network promotes conformation state-dependent eviction of the Nas6 assembly chaperone from nascent 26S proteasomes. Cell Rep 26(2):483–495 e485. https://doi.org/10.1016/j.celrep.2018.12.042
Nencioni A, Garuti A, Schwarzenberg K, Cirmena G, Dal Bello G, Rocco I, Barbieri E, Brossart P, Patrone F, Ballestrero A (2006) Proteasome inhibitor-induced apoptosis in human monocyte-derived dendritic cells. Eur J Immunol 36(3):681–689. https://doi.org/10.1002/eji.200535298
Nickell S, Beck F, Scheres SH, Korinek A, Forster F, Lasker K, Mihalache O, Sun N, Nagy I, Sali A, Plitzko JM, Carazo JM, Mann M, Baumeister W (2009) Insights into the molecular architecture of the 26S proteasome. Proc Natl Acad Sci U S A 106(29):11943–11947. https://doi.org/10.1073/pnas.0905081106
Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13(12):1211–1218. https://doi.org/10.1111/j.1365-2443.2008.01238.x
Noda NN, Ohsumi Y, Inagaki F (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 584(7):1379–1385. https://doi.org/10.1016/j.febslet.2010.01.018
Nussbaum AK, Dick TP, Keilholz W, Schirle M, Stevanovic S, Dietz K, Heinemeyer W, Groll M, Wolf DH, Huber R, Rammensee HG, Schild H (1998) Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc Natl Acad Sci U S A 95(21):12504–12509. https://doi.org/10.1073/pnas.95.21.12504
Ogura T, Wilkinson AJ (2001) AAA + superfamily ATPases: common structure–diverse function. Genes Cells 6(7):575–597
Oh E, Akopian D, Rape M (2018) Principles of ubiquitin-dependent signaling. Annu Rev Cell Dev Biol 34:137–162. https://doi.org/10.1146/annurev-cellbio-100617-062802
Oh S, Hong HS, Hwang E, Sim HJ, Lee W, Shin SJ, Mook-Jung I (2005) Amyloid peptide attenuates the proteasome activity in neuronal cells. Mech Ageing Dev 126(12):1292–1299. https://doi.org/10.1016/j.mad.2005.07.006
Ohno A, Jee J, Fujiwara K, Tenno T, Goda N, Tochio H, Kobayashi H, Hiroaki H, Shirakawa M (2005) Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure 13(4):521–532. https://doi.org/10.1016/j.str.2005.01.011
Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2(3):211–216. https://doi.org/10.1038/35056522
Oinonen C, Rouvinen J (2000) Structural comparison of Ntn-hydrolases. Protein Sci 9(12):2329–2337. https://doi.org/10.1110/ps.9.12.2329
Olivares AO, Baker TA, Sauer RT (2018) Mechanical protein unfolding and degradation. Annu Rev Physiol 80:413–429. https://doi.org/10.1146/annurev-physiol-021317-121303
Olszewski MM, Williams C, Dong KC, Martin A (2019) The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome. Commun Biol 2:29. https://doi.org/10.1038/s42003-019-0283-z
Orlowski M (1990) The multicatalytic proteinase complex, a major extralysosomal proteolytic system. Biochemistry 29(45):10289–10297. https://doi.org/10.1021/bi00497a001
Orlowski RZ, Eswara JR, Lafond-Walker A, Grever MR, Orlowski M, Dang CV (1998) Tumor growth inhibition induced in a murine model of human Burkitt’s lymphoma by a proteasome inhibitor. Cancer Res 58(19):4342–4348
Orlowski RZ, Stinchcombe TE, Mitchell BS, Shea TC, Baldwin AS, Stahl S, Adams J, Esseltine DL, Elliott PJ, Pien CS, Guerciolini R, Anderson JK, Depcik-Smith ND, Bhagat R, Lehman MJ, Novick SC, O’Connor OA, Soignet SL (2002) Phase I trial of the proteasome inhibitor PS-341 in patients with refractory hematologic malignancies. J Clin Oncol 20(22):4420–4427. https://doi.org/10.1200/JCO.2002.01.133
Ortega Z, Diaz-Hernandez M, Lucas JJ (2007) Is the ubiquitin-proteasome system impaired in Huntington’s disease? Cell Mol Life Sci 64(17):2245–2257. https://doi.org/10.1007/s00018-007-7222-8
Ortega Z, Lucas JJ (2014) Ubiquitin-proteasome system involvement in Huntington’s disease. Front Mol Neurosci 7:77. https://doi.org/10.3389/fnmol.2014.00077
Ortolan TG, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K (2000) The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol 2(9):601–608. https://doi.org/10.1038/35023547
Osmulski PA, Hochstrasser M, Gaczynska M (2009) A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure 17(8):1137–1147. https://doi.org/10.1016/j.str.2009.06.011
Osorio FG, Soria-Valles C, Santiago-Fernandez O, Bernal T, Mittelbrunn M, Colado E, Rodriguez F, Bonzon-Kulichenko E, Vazquez J, Porta-de-la-Riva M, Ceron J, Fueyo A, Li J, Green AR, Freije JM, Lopez-Otin C (2016) Loss of the proteostasis factor AIRAPL causes myeloid transformation by deregulating IGF-1 signaling. Nat Med 22(1):91–96. https://doi.org/10.1038/nm.4013
Owsianik G, Balzi l L, Ghislain M (2002) Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae. Mol Microbiol 43(5):1295–1308. https://doi.org/10.1046/j.1365-2958.2002.02823.x
Pack CG, Yukii H, Toh-e A, Kudo T, Tsuchiya H, Kaiho A, Sakata E, Murata S, Yokosawa H, Sako Y, Baumeister W, Tanaka K, Saeki Y (2014) Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat Commun 5:3396. https://doi.org/10.1038/ncomms4396
Padmanabhan A, Vuong SA, Hochstrasser M (2016) Assembly of an evolutionarily conserved alternative proteasome isoform in human cells. Cell Rep 14(12):2962–2974. https://doi.org/10.1016/j.celrep.2016.02.068
Pagan J, Seto T, Pagano M, Cittadini A (2013) Role of the ubiquitin proteasome system in the heart. Circ Res 112(7):1046–1058. https://doi.org/10.1161/CIRCRESAHA.112.300521
Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M (1995) Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269(5224):682–685. https://doi.org/10.1126/science.7624798
Pamnani V, Tamura T, Lupas A, Peters J, Cejka Z, Ashraf W, Baumeister W (1997) Cloning, sequencing and expression of VAT, a CDC48/p97 ATPase homologue from the archaeon Thermoplasma acidophilum. FEBS Lett 404(2–3):263–268. https://doi.org/10.1016/s0014-5793(97)00138-5
Pante N, Kann M (2002) Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell 13(2):425–434. https://doi.org/10.1091/mbc.01-06-0308
Papadopoulos C, Kirchner P, Bug M, Grum D, Koerver L, Schulze N, Poehler R, Dressler A, Fengler S, Arhzaouy K, Lux V, Ehrmann M, Weihl CC, Meyer H (2017) VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J 36(2):135–150. https://doi.org/10.15252/embj.201695148
Park S, Kim W, Tian G, Gygi SP, Finley D (2011) Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. J Biol Chem 286(42):36652–36666. https://doi.org/10.1074/jbc.M111.285924
Park S, Li X, Kim HM, Singh CR, Tian G, Hoyt MA, Lovell S, Battaile KP, Zolkiewski M, Coffino P, Roelofs J, Cheng Y, Finley D (2013) Reconfiguration of the proteasome during chaperone-mediated assembly. Nature 497(7450):512–516. https://doi.org/10.1038/nature12123
Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D (2009) Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 459(7248):866–870. https://doi.org/10.1038/nature08065
Park S, Tian G, Roelofs J, Finley D (2010) Assembly manual for the proteasome regulatory particle: the first draft. Biochem Soc Trans 38(Pt 1):6–13. https://doi.org/10.1042/BST0380006
Parzych KR, Klionsky DJ (2019) Vacuolar hydrolysis and efflux: current knowledge and unanswered questions. Autophagy 15(2):212–227. https://doi.org/10.1080/15548627.2018.1545821
Patel SK, Indig FE, Olivieri N, Levine ND, Latterich M (1998) Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell 92(5):611–620. https://doi.org/10.1016/s0092-8674(00)81129-0
Pathare GR, Nagy I, Bohn S, Unverdorben P, Hubert A, Korner R, Nickell S, Lasker K, Sali A, Tamura T, Nishioka T, Forster F, Baumeister W, Bracher A (2012) The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci U S A 109(1):149–154. https://doi.org/10.1073/pnas.1117648108
Pathare GR, Nagy I, Sledz P, Anderson DJ, Zhou HJ, Pardon E, Steyaert J, Forster F, Bracher A, Baumeister W (2014) Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc Natl Acad Sci U S A 111(8):2984–2989. https://doi.org/10.1073/pnas.1400546111
Perez VI, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, Andziak B, Yang T, Edrey Y, Friguet B, Ward W, Richardson A, Chaudhuri A (2009) Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci U S A 106(9):3059–3064. https://doi.org/10.1073/pnas.0809620106
Peters JM (2006) The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 7(9):644–656. https://doi.org/10.1038/nrm1988
Peters JM, Cejka Z, Harris JR, Kleinschmidt JA, Baumeister W (1993) Structural features of the 26 S proteasome complex. J Mol Biol 234(4):932–937. https://doi.org/10.1006/jmbi.1993.1646
Peters LZ, Karmon O, David-Kadoch G, Hazan R, Yu T, Glickman MH, Ben-Aroya S (2015) The protein quality control machinery regulates its misassembled proteasome subunits. PLoS Genet 11(4):e1005178. https://doi.org/10.1371/journal.pgen.1005178
Peters LZ, Karmon O, Miodownik S, Ben-Aroya S (2016) Proteasome storage granules are transiently associated with the insoluble protein deposit in Saccharomyces cerevisiae. J Cell Sci 129(6):1190–1197. https://doi.org/10.1242/jcs.179648
Peth A, Besche HC, Goldberg AL (2009) Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell 36(5):794–804. https://doi.org/10.1016/j.molcel.2009.11.015
Peth A, Kukushkin N, Bosse M, Goldberg AL (2013a) Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J Biol Chem 288(11):7781–7790. https://doi.org/10.1074/jbc.M112.441907
Peth A, Nathan JA, Goldberg AL (2013b) The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J Biol Chem 288(40):29215–29222. https://doi.org/10.1074/jbc.M113.482570
Peth A, Uchiki T, Goldberg AL (2010) ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol Cell 40(4):671–681. https://doi.org/10.1016/j.molcel.2010.11.002
Pickart CM (2000) Ubiquitin in chains. Trends Biochem Sci 25(11):544–548. https://doi.org/10.1016/s0968-0004(00)01681-9
Piwko W, Jentsch S (2006) Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat Struct Mol Biol 13(8):691–697. https://doi.org/10.1038/nsmb1122
Powell SR, Herrmann J, Lerman A, Patterson C, Wang X (2012) The ubiquitin-proteasome system and cardiovascular disease. Prog Mol Biol Transl Sci 109:295–346. https://doi.org/10.1016/B978-0-12-397863-9.00009-2
Powell SR, Samuel SM, Wang P, Divald A, Thirunavukkarasu M, Koneru S, Wang X, Maulik N (2008) Upregulation of myocardial 11S-activated proteasome in experimental hyperglycemia. J Mol Cell Cardiol 44(3):618–621. https://doi.org/10.1016/j.yjmcc.2007.12.009
Prakash S, Inobe T, Hatch AJ, Matouschek A (2009) Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol 5(1):29–36. https://doi.org/10.1038/nchembio.130
Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A (2004) An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol 11(9):830–837. https://doi.org/10.1038/nsmb814
Puchades C, Ding B, Song A, Wiseman RL, Lander GC, Glynn SE (2019) Unique structural features of the mitochondrial AAA + Protease AFG3L2 reveal the molecular basis for activity in health and disease. Mol Cell. https://doi.org/10.1016/j.molcel.2019.06.016
Puchades C, Rampello AJ, Shin M, Giuliano CJ, Wiseman RL, Glynn SE, Lander GC (2017) Structure of the mitochondrial inner membrane AAA + protease YME1 gives insight into substrate processing. Science 358(6363). https://doi.org/10.1126/science.aao0464
Puchades C, Sandate CR, Lander GC (2020) The molecular principles governing the activity and functional diversity of AAA + proteins. Nat Rev Mol Cell Biol 21(1):43–58. https://doi.org/10.1038/s41580-019-0183-6
Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, Zhong B, Shu HB (2014) RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog 10(9):e1004358. https://doi.org/10.1371/journal.ppat.1004358
Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL (2006) hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J 25(24):5742–5753. https://doi.org/10.1038/sj.emboj.7601450
Raasi S, Pickart CM (2003) Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem 278(11):8951–8959. https://doi.org/10.1074/jbc.m212841200
Raasi S, Varadan R, Fushman D, Pickart CM (2005) Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol 12(8):708–714. https://doi.org/10.1038/nsmb962
Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S (2002) AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol 22(2):626–634. https://doi.org/10.1128/mcb.22.2.626-634.2002
Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y (2008) Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell 30(3):360–368. https://doi.org/10.1016/j.molcel.2008.03.004
Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ (2010) Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell 38(1):17–28. https://doi.org/10.1016/j.molcel.2010.02.029
Rahighi S, Braunstein I, Ternette N, Kessler B, Kawasaki M, Kato R, Matsui T, Weiss TM, Stanhill A, Wakatsuki S (2016) Selective binding of AIRAPL Tandem UIMs to Lys48-linked tri-ubiquitin chains. Structure 24(3):412–422. https://doi.org/10.1016/j.str.2015.12.017
Ramachandran KV, Fu JM, Schaffer TB, Na CH, Delannoy M, Margolis SS (2018) Activity-dependent degradation of the nascentome by the neuronal membrane proteasome. Mol Cell 71(1):169–177 e166. https://doi.org/10.1016/j.molcel.2018.06.013
Ramachandran KV, Margolis SS (2017) A mammalian nervous-system-specific plasma membrane proteasome complex that modulates neuronal function. Nat Struct Mol Biol 24(4):419–430. https://doi.org/10.1038/nsmb.3389
Ramadan K, Bruderer R, Spiga FM, Popp O, Baur T, Gotta M, Meyer HH (2007) Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature 450(7173):1258–1262. https://doi.org/10.1038/nature06388
Ramanathan HN, Ye Y (2012) Cellular strategies for making monoubiquitin signals. Crit Rev Biochem Mol Biol 47(1):17–28. https://doi.org/10.3109/10409238.2011.620943
Ramos PC, Hockendorff J, Johnson ES, Varshavsky A, Dohmen RJ (1998) Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 92(4):489–499. https://doi.org/10.1016/s0092-8674(00)80942-3
Realini C, Dubiel W, Pratt G, Ferrell K, Rechsteiner M (1994) Molecular cloning and expression of a gamma-interferon-inducible activator of the multicatalytic protease. J Biol Chem 269(32):20727–20732
Rechsteiner M, Hill CP (2005) Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol 15(1):27–33. https://doi.org/10.1016/j.tcb.2004.11.003
Reggiori F, Klionsky DJ (2013) Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194(2):341–361. https://doi.org/10.1534/genetics.112.149013
Reits EA, Benham AM, Plougastel B, Neefjes J, Trowsdale J (1997) Dynamics of proteasome distribution in living cells. EMBO J 16(20):6087–6094. https://doi.org/10.1093/emboj/16.20.6087
Religa TL, Sprangers R, Kay LE (2010) Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science 328(5974):98–102. https://doi.org/10.1126/science.1184991
Remondelli P, Renna M (2017) The endoplasmic reticulum unfolded protein response in neurodegenerative disorders and its potential therapeutic significance. Front Mol Neurosci 10:187. https://doi.org/10.3389/fnmol.2017.00187
Renaud JP, Chari A, Ciferri C, Liu WT, Remigy HW, Stark H, Wiesmann C (2018) Cryo-EM in drug discovery: achievements, limitations and prospects. Nat Rev Drug Discov 17(7):471–492. https://doi.org/10.1038/nrd.2018.77
Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S (2005) A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120(1):73–84. https://doi.org/10.1016/j.cell.2004.11.013
Ricoult SJ, Manning BD (2013) The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep 14(3):242–251. https://doi.org/10.1038/embor.2013.5
Riedinger C, Boehringer J, Trempe JF, Lowe ED, Brown NR, Gehring K, Noble ME, Gordon C, Endicott JA (2010) Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J Biol Chem 285(44):33992–34003. https://doi.org/10.1074/jbc.M110.134510
Ripstein ZA, Huang R, Augustyniak R, Kay LE, Rubinstein JL (2017) Structure of a AAA + unfoldase in the process of unfolding substrate. Elife 6. https://doi.org/10.7554/elife.25754
Ripstein ZA, Vahidi S, Houry WA, Rubinstein JL, Kay LE (2020) A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery. Elife 9. https://doi.org/10.7554/elife.52158
Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78(5):761–771. https://doi.org/10.1016/s0092-8674(94)90462-6
Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, Finley D (2009) Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459(7248):861–865. https://doi.org/10.1038/nature08063
Roeten MSF, Cloos J, Jansen G (2018) Positioning of proteasome inhibitors in therapy of solid malignancies. Cancer Chemother Pharmacol 81(2):227–243. https://doi.org/10.1007/s00280-017-3489-0
Rogov V, Dotsch V, Johansen T, Kirkin V (2014) Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 53(2):167–178. https://doi.org/10.1016/j.molcel.2013.12.014
Rogov VV, Stolz A, Ravichandran AC, Rios-Szwed DO, Suzuki H, Kniss A, Lohr F, Wakatsuki S, Dotsch V, Dikic I, Dobson RC, McEwan DG (2017) Structural and functional analysis of the GABARAP interaction motif (GIM). EMBO Rep 18(8):1382–1396. https://doi.org/10.15252/embr.201643587
Romano R, Palamaro L, Fusco A, Giardino G, Gallo V, Del Vecchio L, Pignata C (2013) FOXN1: a master regulator gene of thymic epithelial development program. Front Immunol 4:187. https://doi.org/10.3389/fimmu.2013.00187
Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH (2008) The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol 15(6):573–580. https://doi.org/10.1038/nsmb.1427
Rousseau A, Bertolotti A (2016) An evolutionarily conserved pathway controls proteasome homeostasis. Nature 536(7615):184–189. https://doi.org/10.1038/nature18943
Rousseau A, Bertolotti A (2018) Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol 19(11):697–712. https://doi.org/10.1038/s41580-018-0040-z
Rubinsztein DC (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443(7113):780–786. https://doi.org/10.1038/nature05291
Ruggeri B, Miknyoczki S, Dorsey B, Hui AM (2009) The development and pharmacology of proteasome inhibitors for the management and treatment of cancer. Adv Pharmacol 57:91–135. https://doi.org/10.1016/S1054-3589(08)57003-7
Ruschak AM, Religa TL, Breuer S, Witt S, Kay LE (2010) The proteasome antechamber maintains substrates in an unfolded state. Nature 467(7317):868–871. https://doi.org/10.1038/nature09444
Russell SJ, Steger KA, Johnston SA (1999) Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J Biol Chem 274(31):21943–21952. https://doi.org/10.1074/jbc.274.31.21943
Sadre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP (2010) Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol Cell 37(5):728–735. https://doi.org/10.1016/j.molcel.2010.02.002
Saeki Y (2017) Ubiquitin recognition by the proteasome. J Biochem 161(2):113–124. https://doi.org/10.1093/jb/mvw091
Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, Tanaka K (2009a) Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J 28(4):359–371. https://doi.org/10.1038/emboj.2008.305
Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K (2009b) Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell 137(5):900–913. https://doi.org/10.1016/j.cell.2009.05.005
Saez I, Vilchez D (2014) The mechanistic links between proteasome activity, aging and age-related diseases. Curr Genomics 15(1):38–51. https://doi.org/10.2174/138920291501140306113344
Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H, Sixma TK (2015) Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell 57(5):887–900. https://doi.org/10.1016/j.molcel.2014.12.039
Sakata E, Bohn S, Mihalache O, Kiss P, Beck F, Nagy I, Nickell S, Tanaka K, Saeki Y, Forster F, Baumeister W (2012) Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc Natl Acad Sci U S A 109(5):1479–1484. https://doi.org/10.1073/pnas.1119394109
Sakata E, Yamaguchi Y, Kurimoto E, Kikuchi J, Yokoyama S, Yamada S, Kawahara H, Yokosawa H, Hattori N, Mizuno Y, Tanaka K, Kato K (2003) Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep 4(3):301–306. https://doi.org/10.1038/sj.embor.embor764
Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky AJ, Podlutskaya N, Richardson A, Austad SN, Chaudhuri AR (2009) The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. FASEB J 23(7):2317–2326. https://doi.org/10.1096/fj.08-122523
Samant RS, Livingston CM, Sontag EM, Frydman J (2018) Distinct proteostasis circuits cooperate in nuclear and cytoplasmic protein quality control. Nature 563(7731):407–411. https://doi.org/10.1038/s41586-018-0678-x
Sandstrom A, Mitchell PS, Goers L, Mu EW, Lesser CF, Vance RE (2019) Functional degradation: a mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364(6435). https://doi.org/10.1126/science.aau1330
Sato Y, Goto E, Shibata Y, Kubota Y, Yamagata A, Goto-Ito S, Kubota K, Inoue J, Takekawa M, Tokunaga F, Fukai S (2015) Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat Struct Mol Biol 22(3):222–229. https://doi.org/10.1038/nsmb.2970
Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S (2008) Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455(7211):358–362. https://doi.org/10.1038/nature07254
Satoh K, Sasajima H, Nyoumura KI, Yokosawa H, Sawada H (2001) Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry 40(2):314–319. https://doi.org/10.1021/bi001815n
Sauer RT, Baker TA (2011) AAA + proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem 80:587–612. https://doi.org/10.1146/annurev-biochem-060408-172623
Savulescu AF, Glickman MH (2011) Proteasome activator 200: the heat is on. Mol Cell Proteomics 10(5):R110 006890. https://doi.org/10.1074/mcp.r110.006890
Savulescu AF, Shorer H, Kleifeld O, Cohen I, Gruber R, Glickman MH, Harel A (2011) Nuclear import of an intact preassembled proteasome particle. Mol Biol Cell 22(6):880–891. https://doi.org/10.1091/mbc.E10-07-0595
Scapin G, Potter CS, Carragher B (2018) Cryo-EM for small molecules discovery, design, understanding, and application. Cell Chem Biol 25(11):1318–1325. https://doi.org/10.1016/j.chembiol.2018.07.006
Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391(6668):715–718. https://doi.org/10.1038/35661
Scherer DC, Brockman JA, Chen Z, Maniatis T, Ballard DW (1995) Signal-induced degradation of I kappa B alpha requires site-specific ubiquitination. Proc Natl Acad Sci U S A 92(24):11259–11263. https://doi.org/10.1073/pnas.92.24.11259
Schlossarek S, Carrier L (2011) The ubiquitin-proteasome system in cardiomyopathies. Curr Opin Cardiol 26(3):190–195. https://doi.org/10.1097/HCO.0b013e32834598fe
Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D (2005) The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol 12(4):294–303. https://doi.org/10.1038/nsmb914
Schmidtke G, Kraft R, Kostka S, Henklein P, Frommel C, Lowe J, Huber R, Kloetzel PM, Schmidt M (1996) Analysis of mammalian 20S proteasome biogenesis: the maturation of beta-subunits is an ordered two-step mechanism involving autocatalysis. EMBO J 15(24):6887–6898
Schrader J, Henneberg F, Mata RA, Tittmann K, Schneider TR, Stark H, Bourenkov G, Chari A (2016) The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design. Science 353(6299):594–598. https://doi.org/10.1126/science.aaf8993
Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453(7194):548–552. https://doi.org/10.1038/nature06924
Schweitzer A, Aufderheide A, Rudack T, Beck F, Pfeifer G, Plitzko JM, Sakata E, Schulten K, Forster F, Baumeister W (2016) Structure of the human 26S proteasome at a resolution of 3.9 A. Proc Natl Acad Sci U S A 113(28):7816–7821. https://doi.org/10.1073/pnas.1608050113
Schwertman P, Bekker-Jensen S, Mailand N (2016) Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Biol 17(6):379–394. https://doi.org/10.1038/nrm.2016.58
Seemuller E, Lupas A, Baumeister W (1996) Autocatalytic processing of the 20S proteasome. Nature 382(6590):468–471. https://doi.org/10.1038/382468a0
Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W (1995a) Proteasome from Thermoplasma acidophilum: a threonine protease. Science 268(5210):579–582. https://doi.org/10.1126/science.7725107
Seemuller E, Lupas A, Zuhl F, Zwickl P, Baumeister W (1995b) The proteasome from Thermoplasma acidophilum is neither a cysteine nor a serine protease. FEBS Lett 359(2–3):173–178. https://doi.org/10.1016/0014-5793(95)00036-9
Selkoe DJ (2003) Folding proteins in fatal ways. Nature 426(6968):900–904. https://doi.org/10.1038/nature02264
Shabek N, Herman-Bachinsky Y, Buchsbaum S, Lewinson O, Haj-Yahya M, Hejjaoui M, Lashuel HA, Sommer T, Brik A, Ciechanover A (2012) The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell 48(1):87–97. https://doi.org/10.1016/j.molcel.2012.07.011
Shah SA, Potter MW, McDade TP, Ricciardi R, Perugini RA, Elliott PJ, Adams J, Callery MP (2001) 26S proteasome inhibition induces apoptosis and limits growth of human pancreatic cancer. J Cell Biochem 82(1):110–122. https://doi.org/10.1002/jcb.1150
Sharif H, Wang L, Wang WL, Magupalli VG, Andreeva L, Qiao Q, Hauenstein AV, Wu Z, Nunez G, Mao Y, Wu H (2019) Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570(7761):338–343. https://doi.org/10.1038/s41586-019-1295-z
Sharipo A, Imreh M, Leonchiks A, Imreh S, Masucci MG (1998) A minimal glycine-alanine repeat prevents the interaction of ubiquitinated I kappaB alpha with the proteasome: a new mechanism for selective inhibition of proteolysis. Nat Med 4(8):939–944. https://doi.org/10.1038/nm0898-939
Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, Shi Y, Zhang N, de Poot SA, Tuebing F, Sun S, Vannoy J, Tarasov SG, Engen JR, Finley D, Walters KJ (2016) Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351(6275). https://doi.org/10.1126/science.aad9421
Shibahara T, Kawasaki H, Hirano H (2002) Identification of the 19S regulatory particle subunits from the rice 26S proteasome. Eur J Biochem 269(5):1474–1483. https://doi.org/10.1046/j.1432-1033.2002.02792.x
Shimizu Y, Taraborrelli L, Walczak H (2015) Linear ubiquitination in immunity. Immunol Rev 266(1):190–207. https://doi.org/10.1111/imr.12309
Shirozu R, Yashiroda H, Murata S (2015) Identification of minimum Rpn4-responsive elements in genes related to proteasome functions. FEBS Lett 589(8):933–940. https://doi.org/10.1016/j.febslet.2015.02.025
Shrestha RK, Ronau JA, Davies CW, Guenette RG, Strieter ER, Paul LN, Das C (2014) Insights into the mechanism of deubiquitination by JAMM deubiquitinases from cocrystal structures of the enzyme with the substrate and product. Biochemistry 53(19):3199–3217. https://doi.org/10.1021/bi5003162
Shukla SK, Rafiq K (2019) Proteasome biology and therapeutics in cardiac diseases. Transl Res 205:64–76. https://doi.org/10.1016/j.trsl.2018.09.003
Sijts A, Sun Y, Janek K, Kral S, Paschen A, Schadendorf D, Kloetzel PM (2002) The role of the proteasome activator PA28 in MHC class I antigen processing. Mol Immunol 39(3–4):165–169. https://doi.org/10.1016/s0161-5890(02)00099-8
Sijts EJ, Kloetzel PM (2011) The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell Mol Life Sci 68(9):1491–1502. https://doi.org/10.1007/s00018-011-0657-y
Silva GM, Finley D, Vogel C (2015) K63 polyubiquitination is a new modulator of the oxidative stress response. Nat Struct Mol Biol 22(2):116–123. https://doi.org/10.1038/nsmb.2955
Silva GM, Netto LE, Simoes V, Santos LF, Gozzo FC, Demasi MA, Oliveira CL, Bicev RN, Klitzke CF, Sogayar MC, Demasi M (2012) Redox control of 20S proteasome gating. Antioxid Redox Signal 16(11):1183–1194. https://doi.org/10.1089/ars.2011.4210
Skrott Z, Mistrik M, Andersen KK, Friis S, Majera D, Gursky J, Ozdian T, Bartkova J, Turi Z, Moudry P, Kraus M, Michalova M, Vaclavkova J, Dzubak P, Vrobel I, Pouckova P, Sedlacek J, Miklovicova A, Kutt A, Li J, Mattova J, Driessen C, Dou QP, Olsen J, Hajduch M, Cvek B, Deshaies RJ, Bartek J (2017) Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 552(7684):194–199. https://doi.org/10.1038/nature25016
Sledz P, Unverdorben P, Beck F, Pfeifer G, Schweitzer A, Forster F, Baumeister W (2013) Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci U S A 110(18):7264–7269. https://doi.org/10.1073/pnas.1305782110
Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L (2001) Distinct functional surface regions on ubiquitin. J Biol Chem 276(32):30483–30489. https://doi.org/10.1074/jbc.M103248200
Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS (1996) HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14(6):354–360, 376
Smith DM, Benaroudj N, Goldberg A (2006) Proteasomes and their associated ATPases: a destructive combination. J Struct Biol 156(1):72–83. https://doi.org/10.1016/j.jsb.2006.04.012
Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL (2007) Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell 27(5):731–744. https://doi.org/10.1016/j.molcel.2007.06.033
Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL (2011) ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 144(4):526–538. https://doi.org/10.1016/j.cell.2011.02.005
Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL (2005) ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell 20(5):687–698. https://doi.org/10.1016/j.molcel.2005.10.019
Smith HL, Mallucci GR (2016) The unfolded protein response: mechanisms and therapy of neurodegeneration. Brain 139(Pt 8):2113–2121. https://doi.org/10.1093/brain/aww101
Snoberger A, Brettrager EJ, Smith DM (2018) Conformational switching in the coiled-coil domains of a proteasomal ATPase regulates substrate processing. Nat Commun 9(1):2374. https://doi.org/10.1038/s41467-018-04731-6
Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M (2010) The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev 24(13):1434–1447. https://doi.org/10.1101/gad.1925010
Song Y, Park PMC, Wu L, Ray A, Picaud S, Li D, Wimalasena VK, Du T, Filippakopoulos P, Anderson KC, Qi J, Chauhan D (2019) Development and preclinical validation of a novel covalent ubiquitin receptor Rpn13 degrader in multiple myeloma. Leukemia 33(11):2685–2694. https://doi.org/10.1038/s41375-019-0467-z
Song Y, Ray A, Li S, Das DS, Tai YT, Carrasco RD, Chauhan D, Anderson KC (2016) Targeting proteasome ubiquitin receptor Rpn13 in multiple myeloma. Leukemia 30(9):1877–1886. https://doi.org/10.1038/leu.2016.97
Specht S, Miller SB, Mogk A, Bukau B (2011) Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol 195(4):617–629. https://doi.org/10.1083/jcb.201106037
Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D (2000) Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102(1):67–76. https://doi.org/10.1016/s0092-8674(00)00011-8
Spencer E, Jiang J, Chen ZJ (1999) Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev 13(3):284–294. https://doi.org/10.1101/gad.13.3.284
Sprangers R, Kay LE (2007) Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 445(7128):618–622. https://doi.org/10.1038/nature05512
Sprangers R, Li X, Mao X, Rubinstein JL, Schimmer AD, Kay LE (2008) TROSY-based NMR evidence for a novel class of 20S proteasome inhibitors. Biochemistry 47(26):6727–6734. https://doi.org/10.1021/bi8005913
Stadtmueller BM, Ferrell K, Whitby FG, Heroux A, Robinson H, Myszka DG, Hill CP (2010) Structural models for interactions between the 20S proteasome and its PAN/19S activators. J Biol Chem 285(1):13–17. https://doi.org/10.1074/jbc.C109.070425
Stadtmueller BM, Hill CP (2011) Proteasome activators. Mol Cell 41(1):8–19. https://doi.org/10.1016/j.molcel.2010.12.020
Stadtmueller BM, Kish-Trier E, Ferrell K, Petersen CN, Robinson H, Myszka DG, Eckert DM, Formosa T, Hill CP (2012) Structure of a proteasome Pba1-Pba2 complex: implications for proteasome assembly, activation, and biological function. J Biol Chem 287(44):37371–37382. https://doi.org/10.1074/jbc.M112.367003
Stanhill A, Haynes CM, Zhang Y, Min G, Steele MC, Kalinina J, Martinez E, Pickart CM, Kong XP, Ron D (2006) An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol Cell 23(6):875–885. https://doi.org/10.1016/j.molcel.2006.07.023
Stein A, Ruggiano A, Carvalho P, Rapoport TA (2014) Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell 158(6):1375–1388. https://doi.org/10.1016/j.cell.2014.07.050
Stolz A, Hilt W, Buchberger A, Wolf DH (2011) Cdc48: a power machine in protein degradation. Trends Biochem Sci 36(10):515–523. https://doi.org/10.1016/j.tibs.2011.06.001
Sun S, Liu S, Zhang Z, Zeng W, Sun C, Tao T, Lin X, Feng XH (2017) Phosphatase UBLCP1 controls proteasome assembly. Open Biol 7(5). https://doi.org/10.1098/rsob.170042
Sun XM, Butterworth M, MacFarlane M, Dubiel W, Ciechanover A, Cohen GM (2004) Caspase activation inhibits proteasome function during apoptosis. Mol Cell 14(1):81–93. https://doi.org/10.1016/s1097-2765(04)00156-x
Tai HC, Schuman EM (2008) Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci 9(11):826–838. https://doi.org/10.1038/nrn2499
Takagi K, Kim S, Yukii H, Ueno M, Morishita R, Endo Y, Kato K, Tanaka K, Saeki Y, Mizushima T (2012) Structural basis for specific recognition of Rpt1p, an ATPase subunit of 26 S proteasome, by proteasome-dedicated chaperone Hsm3p. J Biol Chem 287(15):12172–12182. https://doi.org/10.1074/jbc.M112.345876
Takeda K, Yanagida M (2005) Regulation of nuclear proteasome by Rhp6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell 122(3):393–405. https://doi.org/10.1016/j.cell.2005.05.023
Takeuchi J, Chen H, Coffino P (2007) Proteasome substrate degradation requires association plus extended peptide. EMBO J 26(1):123–131. https://doi.org/10.1038/sj.emboj.7601476
Tanahashi N, Murakami Y, Minami Y, Shimbara N, Hendil KB, Tanaka K (2000) Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J Biol Chem 275(19):14336–14345. https://doi.org/10.1074/jbc.275.19.14336
Tanaka K (2009) The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci 85(1):12–36. https://doi.org/10.2183/pjab.85.12
Tanaka K, Ichihara A (1989) Half-life of proteasomes (multiprotease complexes) in rat liver. Biochem Biophys Res Commun 159(3):1309–1315. https://doi.org/10.1016/0006-291x(89)92253-5
Tanaka K, Waxman L, Goldberg AL (1983) ATP serves two distinct roles in protein degradation in reticulocytes, one requiring and one independent of ubiquitin. J Cell Biol 96(6):1580–1585. https://doi.org/10.1083/jcb.96.6.1580
Tanaka K, Yoshimura T, Tamura T, Fujiwara T, Kumatori A, Ichihara A (1990) Possible mechanism of nuclear translocation of proteasomes. FEBS Lett 271(1–2):41–46. https://doi.org/10.1016/0014-5793(90)80367-r
Tang WK, Li D, Li CC, Esser L, Dai R, Guo L, Xia D (2010) A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J 29(13):2217–2229. https://doi.org/10.1038/emboj.2010.104
Tang WK, Xia D (2016) Mutations in the Human AAA(+) chaperone p97 and related diseases. Front Mol Biosci 3:79. https://doi.org/10.3389/fmolb.2016.00079
Taylor EB, Rutter J (2011) Mitochondrial quality control by the ubiquitin-proteasome system. Biochem Soc Trans 39(5):1509–1513. https://doi.org/10.1042/BST0391509
Thompson D, Hakala K, DeMartino GN (2009) Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. J Biol Chem 284(37):24891–24903. https://doi.org/10.1074/jbc.M109.023218
Thomsen ND, Berger JM (2009) Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell 139(3):523–534. https://doi.org/10.1016/j.cell.2009.08.043
Thrower JS, Hoffman L, Rechsteiner M, Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19(1):94–102. https://doi.org/10.1093/emboj/19.1.94
Tian G, Park S, Lee MJ, Huck B, McAllister F, Hill CP, Gygi SP, Finley D (2011) An asymmetric interface between the regulatory and core particles of the proteasome. Nat Struct Mol Biol 18(11):1259–1267. https://doi.org/10.1038/nsmb.2147
Tian L, Holmgren RA, Matouschek A (2005) A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat Struct Mol Biol 12(12):1045–1053. https://doi.org/10.1038/nsmb1018
Tian Z, Zheng H, Li J, Li Y, Su H, Wang X (2012) Genetically induced moderate inhibition of the proteasome in cardiomyocytes exacerbates myocardial ischemia-reperfusion injury in mice. Circ Res 111(5):532–542. https://doi.org/10.1161/CIRCRESAHA.112.270983
Tomaru U, Takahashi S, Ishizu A, Miyatake Y, Gohda A, Suzuki S, Ono A, Ohara J, Baba T, Murata S, Tanaka K, Kasahara M (2012) Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. Am J Pathol 180(3):963–972. https://doi.org/10.1016/j.ajpath.2011.11.012
Tomita T, Hirayama S, Sakurai Y, Ohte Y, Yoshihara H, Saeki Y, Hamazaki J, Murata S (2019) Specific modification of aged proteasomes revealed by tag-exchangeable knock-in mice. Mol Cell Biol 39(1). https://doi.org/10.1128/mcb.00426-18
Tomko RJ Jr, Hochstrasser M (2011) Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Mol Cell 44(6):907–917. https://doi.org/10.1016/j.molcel.2011.11.020
Tomko RJ Jr, Hochstrasser M (2013) Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem 82:415–445. https://doi.org/10.1146/annurev-biochem-060410-150257
Tomko RJ Jr, Hochstrasser M (2014) The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis. Mol Cell 53(3):433–443. https://doi.org/10.1016/j.molcel.2013.12.009
Tomko RJ Jr, Taylor DW, Chen ZA, Wang HW, Rappsilber J, Hochstrasser M (2015) A single alpha helix drives extensive remodeling of the proteasome lid and completion of regulatory particle assembly. Cell 163(2):432–444. https://doi.org/10.1016/j.cell.2015.09.022
Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M (2009) Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol 29(4):1095–1106. https://doi.org/10.1128/MCB.01227-08
Trader DJ, Simanski S, Kodadek T (2015) A reversible and highly selective inhibitor of the proteasomal ubiquitin receptor rpn13 is toxic to multiple myeloma cells. J Am Chem Soc 137(19):6312–6319. https://doi.org/10.1021/jacs.5b02069
Tran HJ, Allen MD, Lowe J, Bycroft M (2003) Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry 42(39):11460–11465. https://doi.org/10.1021/bi035033g
Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101(3):249–258. https://doi.org/10.1016/s0092-8674(00)80835-1
Tsai FT, Hill CP (2020) Same structure, different mechanisms? Elife 9. https://doi.org/10.7554/elife.56501
Tsai YC, Weissman AM (2010) The unfolded protein response, degradation from endoplasmic reticulum and cancer. Genes Cancer 1(7):764–778. https://doi.org/10.1177/1947601910383011
Tseng BP, Green KN, Chan JL, Blurton-Jones M, LaFerla FM (2008) Abeta inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol Aging 29(11):1607–1618. https://doi.org/10.1016/j.neurobiolaging.2007.04.014
Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, Tanaka K, Saeki Y (2017) In Vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 axis contributes to K48-linked chain specificity of the proteasome. Mol Cell 66(4):488–502 e487. https://doi.org/10.1016/j.molcel.2017.04.024
Turakhiya A, Meyer SR, Marincola G, Bohm S, Vanselow JT, Schlosser A, Hofmann K, Buchberger A (2018) ZFAND1 recruits p 97 and the 26S proteasome to promote the clearance of arsenite-induced stress granules. Mol Cell 70(5):906–919 e907. https://doi.org/10.1016/j.molcel.2018.04.021
Twomey EC, Ji Z, Wales TE, Bodnar NO, Ficarro SB, Marto JA, Engen JR, Rapoport TA (2019) Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science 365(6452). https://doi.org/10.1126/science.aax1033
Uddin MM, Ohigashi I, Motosugi R, Nakayama T, Sakata M, Hamazaki J, Nishito Y, Rode I, Tanaka K, Takemoto T, Murata S, Takahama Y (2017) Foxn1-beta5t transcriptional axis controls CD8(+) T-cell production in the thymus. Nat Commun 8:14419. https://doi.org/10.1038/ncomms14419
Uechi H, Hamazaki J, Murata S (2014) Characterization of the testis-specific proteasome subunit alpha4s in mammals. J Biol Chem 289(18):12365–12374. https://doi.org/10.1074/jbc.M114.558866
Ungvari Z, Csiszar A, Sosnowska D, Philipp EE, Campbell CM, McQuary PR, Chow TT, Coelho M, Didier ES, Gelino S, Holmbeck MA, Kim I, Levy E, Sonntag WE, Whitby PW, Austad SN, Ridgway I (2013) Testing predictions of the oxidative stress hypothesis of aging using a novel invertebrate model of longevity: the giant clam (Tridacna derasa). J Gerontol A Biol Sci Med Sci 68(4):359–367. https://doi.org/10.1093/gerona/gls159
Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T (2002) The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure 10(5):609–618
Unverdorben P, Beck F, Sledz P, Schweitzer A, Pfeifer G, Plitzko JM, Baumeister W, Forster F (2014) Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci U S A 111(15):5544–5549. https://doi.org/10.1073/pnas.1403409111
Ustun S, Hafren A, Liu Q, Marshall RS, Minina EA, Bozhkov PV, Vierstra RD, Hofius D (2018) Bacteria exploit autophagy for proteasome degradation and enhanced virulence in plants. Plant Cell 30(3):668–685. https://doi.org/10.1105/tpc.17.00815
Vale RD (2000) AAA proteins. Lords of the ring. J Cell Biol 150(1):F13–19. https://doi.org/10.1083/jcb.150.1.f13
van den Boom J, Meyer H (2018) VCP/p97-mediated unfolding as a principle in protein homeostasis and signaling. Mol Cell 69(2):182–194. https://doi.org/10.1016/j.molcel.2017.10.028
van der Lee R, Lang B, Kruse K, Gsponer J, Sanchez de Groot N, Huynen MA, Matouschek A, Fuxreiter M, Babu MM (2014) Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep 8(6):1832–1844. https://doi.org/10.1016/j.celrep.2014.07.055
van Deventer S, Menendez-Benito V, van Leeuwen F, Neefjes J (2015) N-terminal acetylation and replicative age affect proteasome localization and cell fitness during aging. J Cell Sci 128(1):109–117. https://doi.org/10.1242/jcs.157354
van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD (1996) The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol 16(11):6020–6028. https://doi.org/10.1128/mcb.16.11.6020
Vander Linden RT, Hemmis CW, Schmitt B, Ndoja A, Whitby FG, Robinson H, Cohen RE, Yao T, Hill CP (2015) Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol Cell 57(5):901–911. https://doi.org/10.1016/j.molcel.2015.01.016
Vangala JR, Dudem S, Jain N, Kalivendi SV (2014) Regulation of PSMB5 protein and beta subunits of mammalian proteasome by constitutively activated signal transducer and activator of transcription 3 (STAT3): potential role in bortezomib-mediated anticancer therapy. J Biol Chem 289(18):12612–12622. https://doi.org/10.1074/jbc.M113.542829
Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D (2004) Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem 279(8):7055–7063. https://doi.org/10.1074/jbc.M309184200
Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D (2005) Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell 18(6):687–698. https://doi.org/10.1016/j.molcel.2005.05.013
Vashist S, Ng DT (2004) Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol 165(1):41–52. https://doi.org/10.1083/jcb.200309132
Vekaria PH, Home T, Weir S, Schoenen FJ, Rao R (2016) Targeting p97 to disrupt protein homeostasis in cancer. Front Oncol 6:181. https://doi.org/10.3389/fonc.2016.00181
Velichutina I, Connerly PL, Arendt CS, Li X, Hochstrasser M (2004) Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. EMBO J 23(3):500–510. https://doi.org/10.1038/sj.emboj.7600059
Verhoef LG, Heinen C, Selivanova A, Halff EF, Salomons FA, Dantuma NP (2009) Minimal length requirement for proteasomal degradation of ubiquitin-dependent substrates. FASEB J 23(1):123–133. https://doi.org/10.1096/fj.08-115055
Verma R, Aravind L, Oania R, McDonald WH, Yates JR 3rd, Koonin EV, Deshaies RJ (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298(5593):611–615. https://doi.org/10.1126/science.1075898
Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 11(10):3425–3439. https://doi.org/10.1091/mbc.11.10.3425
Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118(1):99–110. https://doi.org/10.1016/j.cell.2004.06.014
Verma R, Oania RS, Kolawa NJ, Deshaies RJ (2013) Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife 2:e00308. https://doi.org/10.7554/eLife.00308
Vernace VA, Arnaud L, Schmidt-Glenewinkel T, Figueiredo-Pereira ME (2007) Aging perturbs 26S proteasome assembly in Drosophila melanogaster. FASEB J 21(11):2672–2682. https://doi.org/10.1096/fj.06-6751com
VerPlank JJS, Goldberg AL (2017) Regulating protein breakdown through proteasome phosphorylation. Biochem J 474(19):3355–3371. https://doi.org/10.1042/BCJ20160809
VerPlank JJS, Lokireddy S, Zhao J, Goldberg AL (2019) 26S Proteasomes are rapidly activated by diverse hormones and physiological states that raise cAMP and cause Rpn6 phosphorylation. Proc Natl Acad Sci U S A 116(10):4228–4237. https://doi.org/10.1073/pnas.1809254116
Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, Gage FH, Dillin A (2012) Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489(7415):304–308. https://doi.org/10.1038/nature11468
Vilchez D, Saez I, Dillin A (2014) The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun 5:5659. https://doi.org/10.1038/ncomms6659
Vinitsky A, Cardozo C, Sepp-Lorenzino L, Michaud C, Orlowski M (1994) Inhibition of the proteolytic activity of the multicatalytic proteinase complex (proteasome) by substrate-related peptidyl aldehydes. J Biol Chem 269(47):29860–29866
Vinitsky A, Michaud C, Powers JC, Orlowski M (1992) Inhibition of the chymotrypsin-like activity of the pituitary multicatalytic proteinase complex. Biochemistry 31(39):9421–9428. https://doi.org/10.1021/bi00154a014
Voges D, Zwickl P, Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68:1015–1068. https://doi.org/10.1146/annurev.biochem.68.1.1015
Voorhees PM, Orlowski RZ (2006) The proteasome and proteasome inhibitors in cancer therapy. Annu Rev Pharmacol Toxicol 46:189–213. https://doi.org/10.1146/annurev.pharmtox.46.120604.141300
Voutsadakis IA (2017) Proteasome expression and activity in cancer and cancer stem cells. Tumour Biol 39(3):1010428317692248. https://doi.org/10.1177/1010428317692248
Waite KA, De-La Mota-Peynado A, Vontz G, Roelofs J (2016) Starvation induces proteasome autophagy with different pathways for core and regulatory particles. J Biol Chem 291(7):3239–3253. https://doi.org/10.1074/jbc.M115.699124
Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM (2002) Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41(6):1767–1777. https://doi.org/10.1021/bi011892y
Walters KJ, Lech PJ, Goh AM, Wang Q, Howley PM (2003) DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci U S A 100(22):12694–12699. https://doi.org/10.1073/pnas.1634989100
Wang CY, Mayo MW, Baldwin AS Jr (1996) TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science 274(5288):784–787. https://doi.org/10.1126/science.274.5288.784
Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS Jr (1998) NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 281(5383):1680–1683. https://doi.org/10.1126/science.281.5383.1680
Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ (2006) Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington’s disease by ubiquilin. Hum Mol Genet 15(6):1025–1041. https://doi.org/10.1093/hmg/ddl017
Wang H, Monteiro MJ (2007) Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun 360(2):423–427. https://doi.org/10.1016/j.bbrc.2007.06.097
Wang J, Hartling JA, Flanagan JM (1997) The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91 (4):447–456. https://doi.org/10.1016/s0092-8674(00)80431-6
Wang WL, Yu Z, Castillo-Menendez LR, Sodroski J, Mao Y (2019) Robustness of signal detection in cryo-electron microscopy via a bi-objective-function approach. BMC Bioinformatics 20(1):169. https://doi.org/10.1186/s12859-019-2714-8
Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L (2007) Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry 46(11):3553–3565. https://doi.org/10.1021/bi061994u
Wang X, Xu H, Ju D, Xie Y (2008) Disruption of Rpn4-induced proteasome expression in Saccharomyces cerevisiae reduces cell viability under stressed conditions. Genetics 180(4):1945–1953. https://doi.org/10.1534/genetics.108.094524
Wang X, Yen J, Kaiser P, Huang L (2010) Regulation of the 26S proteasome complex during oxidative stress. Sci Signal 3(151):ra88. https://doi.org/10.1126/scisignal.2001232
Wang Z, Yang J, Kirk C, Fang Y, Alsina M, Badros A, Papadopoulos K, Wong A, Woo T, Bomba D, Li J, Infante JR (2013) Clinical pharmacokinetics, metabolism, and drug-drug interaction of carfilzomib. Drug Metab Dispos 41(1):230–237. https://doi.org/10.1124/dmd.112.047662
Wani PS, Rowland MA, Ondracek A, Deeds EJ, Roelofs J (2015) Maturation of the proteasome core particle induces an affinity switch that controls regulatory particle association. Nat Commun 6:6384. https://doi.org/10.1038/ncomms7384
Wani PS, Suppahia A, Capalla X, Ondracek A, Roelofs J (2016) Phosphorylation of the C-terminal tail of proteasome subunit alpha7 is required for binding of the proteasome quality control factor Ecm29. Sci Rep 6:27873. https://doi.org/10.1038/srep27873
Webb AE, Brunet A (2014) FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci 39(4):159–169. https://doi.org/10.1016/j.tibs.2014.02.003
Weberruss MH, Savulescu AF, Jando J, Bissinger T, Harel A, Glickman MH, Enenkel C (2013) Blm10 facilitates nuclear import of proteasome core particles. EMBO J 32(20):2697–2707. https://doi.org/10.1038/emboj.2013.192
Wehmer M, Rudack T, Beck F, Aufderheide A, Pfeifer G, Plitzko JM, Forster F, Schulten K, Baumeister W, Sakata E (2017) Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc Natl Acad Sci U S A 114(6):1305–1310. https://doi.org/10.1073/pnas.1621129114
Wendler P, Ciniawsky S, Kock M, Kube S (2012) Structure and function of the AAA + nucleotide binding pocket. Biochim Biophys Acta 1823(1):2–14. 10.1016/j.bbamcr.2011.06.014
Wendler P, Lehmann A, Janek K, Baumgart S, Enenkel C (2004) The bipartite nuclear localization sequence of Rpn2 is required for nuclear import of proteasomal base complexes via karyopherin alphabeta and proteasome functions. J Biol Chem 279(36):37751–37762. https://doi.org/10.1074/jbc.M403551200
Wenzel T, Eckerskorn C, Lottspeich F, Baumeister W (1994) Existence of a molecular ruler in proteasomes suggested by analysis of degradation products. FEBS Lett 349(2):205–209. https://doi.org/10.1016/0014-5793(94)00665-2
Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP (2000) Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 408(6808):115–120. https://doi.org/10.1038/35040607
Wilk S, Orlowski M (1983) Evidence that pituitary cation-sensitive neutral endopeptidase is a multicatalytic protease complex. J Neurochem 40(3):842–849. https://doi.org/10.1111/j.1471-4159.1983.tb08056.x
Wilkinson KD, Urban MK, Haas AL (1980) Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J Biol Chem 255(16):7529–7532
Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M (2009) Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A 106(43):18213–18218. https://doi.org/10.1073/pnas.0907887106
Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW (1999) The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev 13(3):270–283. https://doi.org/10.1101/gad.13.3.270
Witt E, Zantopf D, Schmidt M, Kraft R, Kloetzel PM, Kruger E (2000) Characterisation of the newly identified human Ump1 homologue POMP and analysis of LMP7(beta 5i) incorporation into 20 S proteasomes. J Mol Biol 301(1):1–9. https://doi.org/10.1006/jmbi.2000.3959
Witt S, Kwon YD, Sharon M, Felderer K, Beuttler M, Robinson CV, Baumeister W, Jap BK (2006) Proteasome assembly triggers a switch required for active-site maturation. Structure 14(7):1179–1188. https://doi.org/10.1016/j.str.2006.05.019
Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, Di Fiore PP, Polo S (2006) Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol 8(11):1246–1254. https://doi.org/10.1038/ncb1484
Wolf DH, Stolz A (2012) The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim Biophys Acta 1823(1):117–124. https://doi.org/10.1016/j.bbamcr.2011.09.002
Worden EJ, Dong KC, Martin A (2017) An AAA motor-driven mechanical switch in Rpn11 controls deubiquitination at the 26S proteasome. Mol Cell 67(5):799–811 e798. https://doi.org/10.1016/j.molcel.2017.07.023
Worden EJ, Padovani C, Martin A (2014) Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol 21(3):220–227. https://doi.org/10.1038/nsmb.2771
Wright KL, White LC, Kelly A, Beck S, Trowsdale J, Ting JP (1995) Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J Exp Med 181(4):1459–1471. https://doi.org/10.1084/jem.181.4.1459
Wu J, Ma YB, Congdon C, Brett B, Chen S, Xu Y, Ouyang Q, Mao Y (2017) Massively parallel unsupervised single-particle cryo-EM data clustering via statistical manifold learning. PLoS ONE 12(8):e0182130. https://doi.org/10.1371/journal.pone.0182130
Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW (2010) UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A 107(4):1355–1360. https://doi.org/10.1073/pnas.0912802107
Wu X, Karin M (2015) Emerging roles of Lys63-linked polyubiquitylation in immune responses. Immunol Rev 266(1):161–174. https://doi.org/10.1111/imr.12310
Wu X, Rapoport TA (2018) Mechanistic insights into ER-associated protein degradation. Curr Opin Cell Biol 53:22–28. https://doi.org/10.1016/j.ceb.2018.04.004
Wu Y, Luo H, Kanaan N, Wu J (2000) The proteasome controls the expression of a proliferation-associated nuclear antigen Ki-67. J Cell Biochem 76(4):596–604
Xia D, Tang WK, Ye Y (2016) Structure and function of the AAA + ATPase p97/Cdc48p. Gene 583(1):64–77. https://doi.org/10.1016/j.gene.2016.02.042
Xie Y, Varshavsky A (2000) Physical association of ubiquitin ligases and the 26S proteasome. Proc Natl Acad Sci U S A 97(6):2497–2502. https://doi.org/10.1073/pnas.060025497
Xie Y, Varshavsky A (2001) RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A 98(6):3056–3061. https://doi.org/10.1073/pnas.071022298
Xie Y, Varshavsky A (2002) UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat Cell Biol 4(12):1003–1007. https://doi.org/10.1038/ncb889
Xu H, Fu J, Ha SW, Ju D, Zheng J, Li L, Xie Y (2012) The CCAAT box-binding transcription factor NF-Y regulates basal expression of human proteasome genes. Biochim Biophys Acta 1823(4):818–825. https://doi.org/10.1016/j.bbamcr.2012.01.002
Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137(1):133–145. https://doi.org/10.1016/j.cell.2009.01.041
Xu S, Peng G, Wang Y, Fang S, Karbowski M (2011) The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol Biol Cell 22(3):291–300. https://doi.org/10.1091/mbc.E10-09-0748
Xu Y, Wu J, Yin CC, Mao Y (2016) Unsupervised Cryo-EM data clustering through adaptively constrained K-means algorithm. PLoS ONE 11(12):e0167765. https://doi.org/10.1371/journal.pone.0167765
Yao T, Cohen RE (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419(6905):403–407. https://doi.org/10.1038/nature01071
Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW (2008) Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell 31(6):909–917. https://doi.org/10.1016/j.molcel.2008.08.027
Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol 8(9):994–1002. https://doi.org/10.1038/ncb1460
Yashiroda H, Mizushima T, Okamoto K, Kameyama T, Hayashi H, Kishimoto T, Niwa S, Kasahara M, Kurimoto E, Sakata E, Takagi K, Suzuki A, Hirano Y, Murata S, Kato K, Yamane T, Tanaka K (2008) Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat Struct Mol Biol 15(3):228–236. https://doi.org/10.1038/nsmb.1386
Yasuda S, Tsuchiya H, Kaiho A, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T, Baumeister W, Fernandez-Busnadiego R, Tanaka K, Saeki Y (2020) Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578(7794):296–300. https://doi.org/10.1038/s41586-020-1982-9
Yau R, Rape M (2016) The increasing complexity of the ubiquitin code. Nat Cell Biol 18(6):579–586. https://doi.org/10.1038/ncb3358
Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez-Martin N, Matsumoto ML, Dixit VM, Rape M (2017) Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell 171(4):918–933 e920. https://doi.org/10.1016/j.cell.2017.09.040
Ye Y, Rape M (2009) Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10(11):755–764. https://doi.org/10.1038/nrm2780
Yedidi RS, Fatehi AK, Enenkel C (2016) Proteasome dynamics between proliferation and quiescence stages of Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol 51(6):497–512. https://doi.org/10.1080/10409238.2016.1230087
Yokoi M, Hanaoka F (2017) Two mammalian homologs of yeast Rad23, HR23A and HR23B, as multifunctional proteins. Gene 597:1–9. https://doi.org/10.1016/j.gene.2016.10.027
Yu H, Kago G, Yellman CM, Matouschek A (2016a) Ubiquitin-like domains can target to the proteasome but proteolysis requires a disordered region. EMBO J 35(14):1522–1536. https://doi.org/10.15252/embj.201593147
Yu H, Lupoli TJ, Kovach A, Meng X, Zhao G, Nathan CF, Li H (2018) ATP hydrolysis-coupled peptide translocation mechanism of Mycobacterium tuberculosis ClpB. Proc Natl Acad Sci U S A 115(41):E9560–E9569. https://doi.org/10.1073/pnas.1810648115
Yu H, Matouschek A (2017) Recognition of Client Proteins by the Proteasome. Annu Rev Biophys 46:149–173. https://doi.org/10.1146/annurev-biophys-070816-033719
Yu H, Singh Gautam AK, Wilmington SR, Wylie D, Martinez-Fonts K, Kago G, Warburton M, Chavali S, Inobe T, Finkelstein IJ, Babu MM, Matouschek A (2016b) Conserved Sequence Preferences Contribute to Substrate Recognition by the Proteasome. J Biol Chem 291(28):14526–14539. https://doi.org/10.1074/jbc.M116.727578
Yu Y, Smith DM, Kim HM, Rodriguez V, Goldberg AL, Cheng Y (2010) Interactions of PAN’s C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J 29(3):692–702. https://doi.org/10.1038/emboj.2009.382
Yun C, Stanhill A, Yang Y, Zhang Y, Haynes CM, Xu CF, Neubert TA, Mor A, Philips MR, Ron D (2008) Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc Natl Acad Sci U S A 105(19):7094–7099. https://doi.org/10.1073/pnas.0707025105
Yu Z, Yu Y, Wang F, Myasnikov AG, Coffino P, Cheng Y (2020) Allosteric coupling between alpha-rings of the 20S proteasome. Nat Commun 11(1):4580. https://doi.org/10.1038/s41467-020-18415-7
Zaiss DM, Standera S, Holzhutter H, Kloetzel P, Sijts AJ (1999) The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett 457(3):333–338. https://doi.org/10.1016/s0014-5793(99)01072-8
Zaiss DM, Standera S, Kloetzel PM, Sijts AJ (2002) PI31 is a modulator of proteasome formation and antigen processing. Proc Natl Acad Sci U S A 99(22):14344–14349. https://doi.org/10.1073/pnas.212257299
Zanelli E, Zhou P, Cao H, Smart MK, David CS (1993) Genomic organization and tissue expression of the mouse proteasome gene Lmp-7. Immunogenetics 38(6):400–407. https://doi.org/10.1007/bf00184520
Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, Glickman MH, Fushman D (2009a) Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell 36(6):1018–1033. https://doi.org/10.1016/j.molcel.2009.11.012
Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y (2009b) Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell 34(4):473–484. https://doi.org/10.1016/j.molcel.2009.04.021
Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE (2003) O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115(6):715–725. https://doi.org/10.1016/s0092-8674(03)00974-7
Zhang F, Wu Z, Zhang P, Tian G, Finley D, Shi Y (2009c) Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell 34(4):485–496. https://doi.org/10.1016/j.molcel.2009.04.022
Zhang L, Chen S, Ruan J, Wu J, Tong AB, Yin Q, Li Y, David L, Lu A, Wang WL, Marks C, Ouyang Q, Zhang X, Mao Y, Wu H (2015) Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350(6259):404–409. https://doi.org/10.1126/science.aac5789
Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, Kang Y, Haririnia A, Storaska AJ, Cole JL, Fushman D, Walters KJ (2009d) Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell 35(3):280–290. https://doi.org/10.1016/j.molcel.2009.06.010
Zhang NY, Jacobson AD, Macfadden A, Liu CW (2011) Ubiquitin chain trimming recycles the substrate binding sites of the 26 S proteasome and promotes degradation of lysine 48-linked polyubiquitin conjugates. J Biol Chem 286(29):25540–25546. https://doi.org/10.1074/jbc.M111.260505
Zhang S, Mao Y (2020) AAA + ATPases in Protein Degradation: Structures, Functions and Mechanisms. Biomolecules 10 (4). https://doi.org/10.3390/biom10040629
Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J, Leonard G, Meyer H, van Heel M, Freemont PS (2000) Structure of the AAA ATPase p97. Mol Cell 6(6):1473–1484
Zhang X, Wigley DB (2008) The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA + proteins. Nat Struct Mol Biol 15(11):1223–1227. https://doi.org/10.1038/nsmb.1501
Zhang Y, Manning BD (2015) mTORC1 signaling activates NRF1 to increase cellular proteasome levels. Cell Cycle 14(13):2011–2017. https://doi.org/10.1080/15384101.2015.1044188
Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, Manning BD (2014) Coordinated regulation of protein synthesis and degradation by mTORC1. Nature 513(7518):440–443. https://doi.org/10.1038/nature13492
Zhang Z, Lv X, Yin WC, Zhang X, Feng J, Wu W, Hui CC, Zhang L, Zhao Y (2013) Ter94 ATPase complex targets k11-linked ubiquitinated ci to proteasomes for partial degradation. Dev Cell 25(6):636–644. https://doi.org/10.1016/j.devcel.2013.05.006
Zhao J, Zhai B, Gygi SP, Goldberg AL (2015) mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci U S A 112(52):15790–15797. https://doi.org/10.1073/pnas.1521919112
Zhao M, Zhang NY, Zurawel A, Hansen KC, Liu CW (2010) Degradation of some polyubiquitinated proteins requires an intrinsic proteasomal binding element in the substrates. J Biol Chem 285(7):4771–4780. https://doi.org/10.1074/jbc.M109.060095
Zhao S, Ulrich HD (2010) Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proc Natl Acad Sci U S A 107(17):7704–7709. https://doi.org/10.1073/pnas.0908764107
Zheng N, Shabek N (2017) Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem 86:129–157. https://doi.org/10.1146/annurev-biochem-060815-014922
Zhu Y, Ouyang Q, Mao Y (2017) A deep convolutional neural network approach to single-particle recognition in cryo-electron microscopy. BMC Bioinformatics 18(1):348. https://doi.org/10.1186/s12859-017-1757-y
Zhu Y, Wang WL, Yu D, Ouyang Q, Lu Y, Mao Y (2018) Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome. Nat Commun 9(1):1360. https://doi.org/10.1038/s41467-018-03785-w
Zong N, Ping P, Lau E, Choi HJ, Ng DC, Meyer D, Fang C, Li H, Wang D, Zelaya IM, Yates JR 3rd, Lam MP (2014) Lysine ubiquitination and acetylation of human cardiac 20S proteasomes. Proteomics Clin Appl 8(7–8):590–594. https://doi.org/10.1002/prca.201400029
Zong WX, Edelstein LC, Chen C, Bash J, Gelinas C (1999) The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappaB that blocks TNFalpha-induced apoptosis. Genes Dev 13(4):382–387. https://doi.org/10.1101/gad.13.4.382
Zuhl F, Seemuller E, Golbik R, Baumeister W (1997) Dissecting the assembly pathway of the 20S proteasome. FEBS Lett 418(1–2):189–194. https://doi.org/10.1016/s0014-5793(97)01370-7
Zuin A, Bichmann A, Isasa M, Puig-Sarries P, Diaz LM, Crosas B (2015) Rpn10 monoubiquitination orchestrates the association of the ubiquilin-type DSK2 receptor with the proteasome. Biochem J 472(3):353–365. https://doi.org/10.1042/BJ20150609
Acknowledgements
The author thanks all members of the Mao Laboratory for helpful discussions and particularly Shitao Zou and Shuwen Zhang for a proofreading of the manuscript, Drs. Alfred Goldberg, Daniel Finley, Marc Kirschner, and Ying Lu for constructive discussions relevant to this work. I apologize for not including many relevant excellent literatures due to our current focus and scope of this review and my limitation in referencing efforts. This work was partly funded by National Natural Science Foundation of China, grant number 1177402 and Natural Science Foundation of Beijing Municipality, grant number Z180016.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this chapter
Cite this chapter
Mao, Y. (2021). Structure, Dynamics and Function of the 26S Proteasome. In: Harris, J.R., Marles-Wright, J. (eds) Macromolecular Protein Complexes III: Structure and Function. Subcellular Biochemistry, vol 96. Springer, Cham. https://doi.org/10.1007/978-3-030-58971-4_1
Download citation
DOI: https://doi.org/10.1007/978-3-030-58971-4_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-58970-7
Online ISBN: 978-3-030-58971-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)