Abstract
While genetics has assisted fisheries management for over 50 years, genetic applications aiming to alleviate or eliminate discards have received little attention. In this chapter, we focus on how genetics can be applied under the EU Landing Obligation, to identify and prevent unwanted catches and to estimate the composition of products made from such catches. Three themes are covered: (i) the genetic identification of bycatch; (ii) the genetic analysis of species composition in nutritional products made from unwanted fish; (iii) the potential of using so-called environmental DNA (DNA shedded from aquatic organisms into the water) to reduce bycatch. For all themes, we introduce and explain the relevant genetic techniques, including data formats and analyses. We present the most significant limitations of the methodologies for their implementation in fisheries and provide examples of their use through relevant case studies. Finally, we discuss the potential future perspectives, with emphasis on the rapid progress in portable and automatic DNA devices, which may revolutionize the use of real-time onsite genetic analyses.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
For more than 50 years, genetics has been assisting fisheries management and marine conservation efforts (Ovenden et al. 2015). During this period, the molecular techniques for studying genetic variation and associated analytical methods have changed tremendously (e.g., Goodwin et al. 2016; Herbert et al. 2003), which in turn have prompted new possibilities in relation to fisheries (Ovenden et al. 2015). Examples of the application of genetics are diverse and cover issues like species identification, analysis of stock structure and food traceability, which are all relevant for conservation, fisheries managers and consumers (reviewed by Ovenden et al. 2015). So far, most uses have focussed on ecosystem management and sustainable harvest of fish resources, while genetic applications aiming to alleviate or eliminate discards have received little attention.
In this chapter, we focus on how genetics can be applied under the EU Landing Obligation, in order to assess and mitigate unwanted catches (e.g., undersized juvenile and low-profit species that will have to be retained and landed under the Landing Obligation) and identify the composition of processed products made from these catches. For all themes, we introduce the relevant genetic techniques, the associated data format and downstream analyses. Moreover, we present the most significant limitations of the techniques, provide examples of their use through relevant case studies and finally discuss potential future perspectives.
2 Genomes, Genes and Genetic Markers
All genetic approaches described in this chapter rely on the analysis of Deoxyribonucleic acid (DNA), which constitutes the genetic code. When populations of organisms are isolated from each other, they follow separate evolutionary trajectories and accumulate genetic differences over time due to the evolutionary forces of mutation, selection and genetic drift (stochastic changes in the frequencies of genetic variants accumulating from generation to generation) (Coyne and Orr 2004; Mayr 1942, 1963). Accordingly, when two sympathetic species have been geographically separated for long enough, their DNA will contain genetic differences, which can be used to unambiguously identify these species using various techniques.
Within a normal animal cell, two different genomes can be found, i.e. the nuclear genome and the mitochondrial genome also referred to as the “mitogenome” (Fig. 20.1). The nuclear genome is located within the cell nucleus in one copy per cell. This genome contains most of the organisms’ genes and is by far the largest, consisting of millions or billions of base pairs. The mitochondrial genome is a much smaller circular genome of around 15–17,000 base pairs in length, and normally includes 37 genes (Ballard and Whitlock 2004). One mitogenome is located in each mitochondrion, i.e. the small organelles responsible for energy production (Saraste 1999), which is also found in the cell. The number of mitochondria varies between cell types, with numbers ranging from ~80 to ~2000 in mammals (Cole 2016). Hence, there are many copies of the mitogenome, but only one nuclear genome within each cell. The high number of mitogenomes per cell is an advantage for many genetic applications, as it is easier to obtain a sufficient amount of intact DNA suitable for downstream applications (Ballard and Whitlock 2004; Galtier et al. 2009). Combined with other attributes like a highly conserved gene content and architecture, which promote easy inter-species comparisons, this feature has made the mitochondrial DNA the most widely used marker for many genetic applications.
3 Species Detection and Bycatch Assignment
3.1 The Technology and Its Applications in Fisheries
With the implementation of the Landing Obligation, many species that previously would have been discarded, e.g. undersized juvenile and low-profit species will now have to be retained and landed. This will challenge the fishing industry, requiring authorities and managers to ensure the correct identification of all landed fish. This information will allow for a more accurate assessment of fisheries’ impacts on species and populations, improving possibilities for evaluating the effect of fishing on the viability of the species or populations of interest (Ovenden et al. 2015). Thus, if fishing has a significant impact on certain species, legal actions can be put in place to reduce bycatch.
Species identification is often based on morphological characters alone; however, in many cases proper species assignment can be problematic. For example, diagnostic characters may not be present across juvenile life stages of some species (McEachran and Musick 1973). Moreover, species may lack morphological characters for easy taxonomic identification because they are badly preserved (or damaged by the fishing gear), are poorly defined (due to limited taxonomic knowledge), or do not exist (cryptic species, i.e. different species that are morphological identical) (Ovenden et al. 2015). Even when good diagnostic morphological characters exist, taxonomic analysis can be difficult, labour intensive and expensive, as it relies on the expertise of highly specialised taxonomists, the number of which is declining worldwide (Hopkins and Freckleton 2002; Wheeler et al. 2004). Hence, alternative ways of conducting species assignment are warranted.
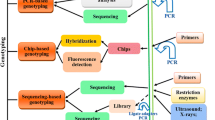
DNA-based identification methods offer fast, standardized and accurate tools for species assignment. The only prerequisites are that the focal species and its sister-species have been described taxonomically, that regions of their DNA have been sequenced and that there is sufficient variation within these sequences to discriminate between target and other taxa. The most commonly used technique is “DNA barcoding” described by Herbert et al. (2003) (for description of technical terms in bold, see text box 20.1). This technique relies on region-specific DNA amplification using a method known as Polymerase Chain Reaction (PCR) and is typically followed by conventional Sanger DNA sequencing. Typically, for animals, the mitochondrial cytochrome oxidase subunit 1 gene (abbreviated COI) is sequenced and compared to a validated sequence reference database (Fig. 20.2a).
Overview of the analytical steps in genetic species-identification either by (a) DNA barcoding or (b) Quantitative PCR (qPCR). In DNA barcoding, the barcoding gene (e.g. COI) is amplified by PCR and the product subsequently sequenced and finally compared to a database for species-match. In qPCR species-specific assays are used. Such assays are initially tested for possible cross-amplification to other species in order to control for false-positive detections. Hence, a positive amplification signal denotes a positive identification for the tested species
The amplified gene segment is approximately 650 base pairs long and was originally chosen based on the level of sequence variation, for most taxa showing only minute genetic variation within the species, while still enabling the discrimination of even closely related species (Hebert et al. 2003). Moreover, several universal PCR assays have been developed rendering it possible to amplify and sequence COI across large taxonomic groups, e.g. fish, vertebrates and invertebrates (Ivanova et al. 2007). However, other gene regions of either mitochondrial or nuclear origin are also used, but to a lesser degree, provided they are diagnostic for the species under investigation. For example, in fish the mitochondrial cytochrome b gene (Cytb) is extensively used as a DNA marker for species identification (Teletchea 2009).
Textbox 20.1 Descriptions of the Molecular Methods Mentioned in This Chapter
PCR
Polymerase Chain Reaction or PCR is a method to amplify a specific region of the DNA, allowing the generation of billions of DNA copies of specific genomic regions. The technique relies on two short DNA sequences, so-called primers, and an enzyme, a polymerase, which copies DNA during repeated cycles of heating and cooling (so-called PCR cycles). Primers only bind to a specific region of the target DNA and serve as starting point for the DNA polymerase. PCR can be conducted with species-specific primers, targeting a single species, or universal primers that amplifies a specific region across taxa. The latter is normally used for DNA barcoding when the identity of the specimen is not known a priori.
PCR assay
The specific combination of primers used in PCR. For qPCR and ddPCR the assay include primers and reporter molecule.
DNA barcoding
DNA barcoding relies on PCR using universal primers that amplifies a specific region, normally the mitochondrial COI gene, across taxa. Following amplification, the region can be purified, sequenced and finally compared to a database in order to establish species identity.
Sanger sequencing
Sanger sequencing is a technique that allows for DNA sequencing of one sequence at the time of up to around 1000 base pairs.
Next generation sequencing
Next generation sequencing or NGS refers to DNA sequencing, which allows individual sequencing of million of sequences in parallel. Many platforms only generate short sequences up to few hundred base pairs.
Meta-barcoding
Meta-barcoding is a method that combines DNA based identification and Next generation Sequencing DNA sequencing. It relies on the use of universal primers, which allows amplification of a specific target region across species groups (e.g. fish) in environmental or other mixed species samples, which subsequently can be sequenced. Output sequences can following be matched to a reference database in order to analyse the number of species present in the sample. Moreover, a measure of relative species composition can be made by counting the number of sequences from each observed species. However, in some cases, the primers may work better for some species than other, which will bias such estimates.
qPCR
Real-time PCR, also called quantitative PCR or qPCR, is a method that allows for analysis of a PCR reaction in real-time, which enables enumeration of DNA copies. qPCR is based on the same approach as PCR but additionally contains a fluorescence region-specific reporter molecule, which allows for visualization of the increase in DNA copies during PCR. A positive detection is based on the accumulation of the fluorescent signal and the cycle threshold (Ct), which is defined as the number of cycles required for the signal to be significantly higher than the background fluorescence level. If a qPCR reaction is analysed together with a dilution series (a standard curve) of a known number of targets, the number of DNA molecules in the reaction can be estimated.
ddPCR
Like in qPCR, droplet digital PCR or ddPCR relies on PCR using region specific primers and a DNA specific reporter. However, differently from qPCR the total PCR reaction is fractionated into thousands of small oil droplets, and PCR is subsequently carried out separately within each droplet. Following PCR each droplet is analysed for positive amplification and number of initial DNA targets. Compared to qPCR, ddPCR provide an absolute count of target DNA copies per sample without the need for running a standard curve and enables more accurate measurements of small fold changes in target DNA amongst samples.
When comparing a sequence from a specimen with unknown taxonomic identity to the database, it is important to make sure that the obtained sequence is of high quality and that the database includes all closely related species. If this is the case, the identity of the specimen can be inferred with very high precision, except in cases where diagnostic variants do not exist. A lack of diagnostic variants can be found e.g. between extremely closely related species and in groups of organisms with low levels of variation in the chosen barcoding region or due to introgression events where the DNA is shared between species because of hybridization. When no close matches of the DNA sequence can be found in the database, the sample likely belongs to an unknown species or one that is not included in the database (Ovenden et al. 2015). Genetics can also be used to assign individuals to populations within species (Nielsen 2016). In these cases, several nuclear genetic markers are normally used in a probabilistic framework, combining the statistical power of individual genetic markers into an overall likelihood of origin. This is necessary, as populations are rarely fixed for specific (diagnostic) gene variants of either mitochondrial or nuclear genes.
3.2 Current Limitations
The accuracy of DNA barcoding is generally high with most species showing distinctive COI sequences (Hebert et al. 2003). For example, among 207 Australian fish species, the identification success rate was 93–98% across groups (Ward et al. 2005), and a 90% accuracy has been documented for North American freshwater fish (April et al. 2011). Nonetheless, the overall precision of the methods critically depends on the reference database used (Ovenden et al. 2015). To assure transparency, sequences should optimally be matched against DNA sequence data from voucher specimens or other curated museum specimens to ensure the reliability of the data (Ratnasingham and Hebert 2007). Currently several public DNA databases exist. GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and the Barcode of Life (http://www.boldsystems.org/index.php/databases) are probably the best known and contain sequences from thousands of species. While the Barcode of Life database consists of several recognized DNA barcoding genes with high quality assurance assessment, Genbank acts as a general repository for genetic data and has therefore a lower quality standard. Due to a general increase in the use of genetics for species identification and taxonomic purposes, as well as the establishment of databases for commercial species, the databases already contain sequences from many species and are constantly expanding. For example, the Barcode of Life database (on the 16th of June 2018) contained 18,843 species of ray-finned fishes. However, the databases remain incomplete, especially for some cryptic, rare and non-commercial species and for geographical regions with high species diversity.
Another limiting factor is the labour expenditure needed for sequencing and analysing of the data. However, other genetic methods can be used for performing species-level assignment that do not involve direct visualization of the genetic code, thereby potentially allowing faster and easier species identification. One such technique is real-time PCR, also known as quantitative PCR (qPCR) (Fig. 20.2b). Quantitative PCR relies on species-specific detection and utilizes a chemistry, which emit light when in contact with DNA from the target organism. The technique allows for simultaneous analysis of one or multiple samples in parallel thereby permitting a fast, sensitive and a potentially cost-efficient alternative to conventional DNA barcoding for routine presence/absence applications. While the risk of false-positives in DNA barcoding both depends on the accuracy of the reference databases and the target region’s ability to discriminate between species, false-positives in qPCR analysis depend on potential cross-amplification to non-target species (the method provides a signal of presence of the species, while it is not there). Hence, for qPCR analysis thorough testing for cross-amplification is needed before an assay can be validated. This can be particularly laborious and expensive for marine species due to the often-large number of related and co-occurring species. Moreover, since a single qPCR assay is specific to a particular species or related group of organisms, qPCR is most useful to identify samples where the number of candidate species can be restricted to a few (for example by using morphological characteristics, see Helyar et al. 2014). In cases where the species identity is completely unknown (e.g. highly damaged or processed individuals) many independent qPCR analyses might be needed to provide a positive detection, which increases the cost significantly. In these cases, DNA barcoding may be more cost effective.
3.3 Case Studies
Skates and rays are particularly vulnerable to overharvesting due to their life history patterns involving slow growth rates, high age of maturity and low fecundity and slow reproductive rates (Holden 1973). Currently, several species are considered threatened and are therefore protected, while others are still targeted or caught as bycatch and sold commercially (STECF 2017) . These include several species that can be difficult to distinguish from protected species based on morphology alone (McEachran and Musick 1973): e.g., the thorny skate (Amblyraja radiata) is protected in the North Sea fisheries (STECF 2017) but can be difficult to distinguish from the thornback ray (Raja clavata), which may be landed (Larsen et al. 2013). Moreover, identification is commonly based on the number of dorsal spines, which in cases where only the ray “wings” are retained, as practiced in some countries (e.g., Denmark), renders species assessment nearly impossible - even by trained specialists. Hence, to monitor rays and skates, genetic tools should be applied. This may especially become relevant with the introduction of the Landing Obligation, as there is a risk that protected species will be landed. Here a standardized genetic approach may be of vital importance for accurate species monitoring of the landed catches, which in turn should eventually lead to better management of the species. Examples of this already exist for other species. For example, albatrosses have suffered high mortality as bycatch in long line fisheries (Burg 2007; Walsh and Edwards 2005). These species are highly mobile, migrating thousands of kilometres between breeding and feeding grounds. As feeding habitat often overlaps between species or populations, and because bycatch carcasses may be highly degraded, it has been difficult to assign individuals to species or populations. Analyses of genetic markers either mitochondrial DNA (Walsh and Edwards 2005) alone or in combination with nuclear markers (Burg 2007) have demonstrated that species, sub-species and even breeding populations could be assigned with high accuracy. This has permitting the impacts of bycatch on specific populations or species to be more accurately estimated.
4 Monitoring Species Composition in Processed Fish Products
4.1 The Technology and Its Applications in Fisheries
Millions of tonnes of fish are thrown back to the sea as bycatch every year (Kelleher 2005). With the introduction of the EU Landing Obligation, all catches of regulated commercial species will have to be landed and counted against quota. This procedure is expected to lead to large amounts of bycatch being landed, which can be utilized in the feed or food industry (Iñarra et al., this volume). Given a low price for unwanted catch (e.g. due to low valued species or individuals), methods for minimizing handling time and costs are likely to be implemented, which will probably lead to storing of catch as mixed or bulk products. One promising approach is the application of fish silage (Viðarsson et al., this volume). Fish silage is a liquid product made from grounded fish in the presence of added acids or lactic-acid-producing bacteria. This product can be pre-processed on board at sea, requires low investments and can be stored at room temperature for an extended period (Arruda et al. 2007). Due the expanding aquaculture industry, especially salmon farming, the demand for fish feed has grown. Accordingly, the price for prime fish silage has increased and is now in a price range where it is a valid alternative to other storage methods like freezing (personal communication, Erling P. Larsen, Senior Executive Consultant, DTU Aqua, Technical University of Denmark). Frozen fish can be used for production of feed (fish meal) but also for food like surimi. However, freezing of low cost fish will reduce the storage capacity for more high-priced catch rendering fish silage an economical and practical alternative. Fish silage can also be produced from the discarded fraction of the primary catch, such as guts, liver and other organs (Arruda et al. 2007), which increases the economic motivation for its use. However, before such products can be introduced to the relevant industry, accurate labelling of species composition according to the food safety standard regulation of the European Union, needs to be ensured (EU Commission Directive 2002/86/EC). As this task cannot be done by visual inspections of bulk products like fish silage, other approaches are needed to analyse and quantify species composition.
Extensive research has been conducted into molecular methods enabling assessment of the species-identity of processed meats. Immunological methods have been used for species detection in meat (Ballin et al. 2009). These methods typically rely on the detection of a species-specific protein, but have now largely been replaced by genetic methods. Targeting DNA compared to proteins has several advantages. First of all, DNA has a higher thermal stability, allowing detection in both processed and raw products. DNA is present in all living cells, regardless of tissue origin and hence easier to detect than proteins that may be located in specific tissues (Lockley and Bardsley 2000). Secondly, the variation in the genetic code between species makes DNA more suitable for species discrimination, especially between closely related species. Finally, PCR-based techniques have lower limits of detection (defined as the lowest concentration of target that can be detected on 95% of occasions) with estimates of 0.001%–1% of the total weight in mixed meat samples (Ballin et al. 2009).
Overall, the methods used for species identification can also detect species presence in processed products. For example, DNA Barcoding, has been used in investigations of adulterated labelling of fish products like fresh or frozen fish fillets, fish fingers, fish cakes and surimi (Helyar et al. 2014; Huxley-Jones et al. 2012; Pepe et al. 2007). However, as conventional Sanger sequencing techniques do not allow for analysis of multiple DNA sequences, this technique only qualifies when used on products originating from a single species. Moreover, in processed samples, shorter and more variable DNA segments may be preferred, as the DNA normally is degraded into short fragments often only few hundred of base pairs long (e.g. Pepe et al. 2007). Quantitative PCR can be used for detection of short species-specific DNA sequences (often < 200 bases) in mixed species products and has been used for example for testing the presence of haddock (Melanogrammus aeglefinus) and cod (Gadus morhua) in products labelled as whitefish and sold from supermarkets in the UK (Helyar et al. 2014). This technique also allows for quantitative estimation of DNA copy numbers when analysed in combination with a standard dilution series of known DNA concentration. It therefore combines fast identification with quantitative applications (Fig. 20.3a). Another well-established PCR-based method is the droplet digital PCR (abbreviated ddPCR) (Fig. 20.3b). This technique builds on the same principles as qPCR but is more precise when used for quantification and does not require a standard curve for quantification of DNA copies. Both qPCR and ddPCR have been used with success to analyse species composition in mixed meat products (e.g., Floren et al. 2015), but so far few studies exists for fish (Bojolly et al. 2017). However, with the increased focus on species identification in mixed fish products and a continuous declining cost of the chemistry used, it is likely to become a more common practice within a few years.
Overview of the methods used for DNA quantification using either mixed species samples or eDNA. Initial DNA extraction is universal for all applications. In qPCR studies, (a) a dilution series (standard curve) with a known number of DNA copies (depicted in black) can be used to estimate the number of copies in the unknown sample (red dot). Here cycle threshold value (Ct) is the PCR cycle where the fluorescence signal becomes significantly higher than the background and can be used to estimate the amount of DNA in the unknown sample. Compared to qPCR, (b) ddPCR provides an absolute count of target DNA copies per sample without the need for running a standard curve enabling more accurate measurements of target DNA content among samples. In meta-barcoding, (c) the gene in focus (e.g. COI) is first amplified in order to up-concentrate the number of DNA copies from a specific group of taxa, e.g. fish. Following this, DNA is sequenced using a Next-generation (NGS) platform generating millions of sequences that subsequently can be sorted, quantified and compared to a data base
4.2 Current Limitations
Quantitative DNA based methods can be used to accurately determine the number of DNA molecules present in a mixed-species product. However, translating this into percentages by weight, which is the normal standard within the food industry, is difficult. For example, the number of mitochondria and hence mitochondrial DNA copies per cell varies across cell types, as well as species (Cole 2016). For this reason, several researchers have advocated for the use of nuclear markers, as they exist in a fixed number of copies per cell (Ballin et al. 2009). However, for products like fish silage generated from whole un-gutted fish, complications may still arise. For example, liver and intestines contain more cells and hence nuclear DNA copies per weight than white muscle. As organ size is not fixed relative to the size of the fish, but varies according to size, age, maturity and species, inaccuracies using nuclear DNA is also expected. A solution may be to use a small fraction of the total catch to estimate species weight composition, which then can be used to estimate a correction factor (Thomas et al. 2016) or to label mixed products in a genome to genome equivalent, instead of weight (Ballin et al. 2009). A final issue with using whole fish, or fish guts, is that DNA from prey items also can be detected. As part of the prey may be other commercial species, which in theory would then be deducted from the quota. However, since prey items normally constitute a minute fraction of the entire product, this risk may be avoided by using a minimum threshold for quota deductions.
Another limitation relates to the fact that both qPCR and ddPCR rely on species-specific detections, which make these techniques expensive for screening products of unknown composition. In such cases, meta-barcoding may be a better alternative. In meta-barcoding, a gene, like COI, is sequenced across one taxon (e.g. fish) using a high-throughput next generation sequencing (NGS) platform. This allows for the sequencing of millions of individual gene sequences, which can be matched to a reference database and counted, and hence combines DNA barcoding with possible quantification (Fig. 20.3c). This method has been used to assess species identity and composition for different kinds of mixed species samples. Meta-barcoding has been used for assessing animal diet by analysing the gut content (e.g., Murray et al. 2011) and has been one of the favoured tools for analysing environmental DNA samples (see next section) (Hansen et al. 2018). However, often only a subset of species can be targeted simultaneously due to variation in the DNA code used for PCR amplification, which moreover can lead to quantification bias if some species amplify better than others during PCR. In addition, as most NGS platforms generate short sequences, the sequenced DNA region will be shorter than for conventional Sanger sequencing. This represents a potential problem, as the shorter sequences contain less variation to discriminate between species. Hence, often sequences cannot be resolved to a species level but only to higher taxonomic levels (Pompanon et al. 2012; Stat et al. 2017).
4.3 Case Study
In order for fish silage to be a valid alternative to frozen fish, several questions need to be addressed. Earlier unpublished studies conducted by DTU Aqua, Technical University of Denmark, have demonstrated that DNA is still present in fish silage after 21 days at both 5 and 20 degrees, as confirmed by amplification and sequencing of the mitochondrial COI gene. However, whether the product also allows accurate quantification of species composition remains unknown. This was investigated in a preliminary study, conducted as part of the Discardless project funded by the European Union. Here specific mitochondrial qPCR assays were developed for cod, haddock and whiting. Subsequently fish silage was made from the same species and DNA samples were extracted over a period of 21 days. Finally, qPCR analysis was undertaken in order to estimate the distribution of DNA copy number among species in the mixture, which then could be compared to the relative difference in weight. The results show that, overall, there is a good correspondence between the input (in weight) of the three species and the number of DNA copies present. The largest difference is found at the first day of the experiment, probably indicating insufficient liquefaction and mixing (Fig. 20.4). After that, the difference decreased significantly, although smaller differences could still be observed until day 21. In general, whiting, which represented the smallest biomass, was overrepresented in the DNA quantification. Similar results have been documented in other studies, but can be improved by calculating a correction factor from a subsample (Thomas et al. 2016). Hence, although more empirical studies are needed, the results hold promise for the application of DNA-based methods for determining the composition of species in silage and other mixed products from bycatch.
Analysed species composition in fish silage made from whiting, haddock and Atlantic cod. The estimates are based on qPCR analysis of the number of species-specific DNA copies. The first column represents the actual biomass while the latter show the qPCR estimates following 1, 7, 14 and 21 days from when the silage was produced
5 Environmental DNA and Pre-assessment of Appropriate Fishing Grounds
5.1 The Technology and Its Applications in Fisheries
Traditional DNA-based monitoring of fisheries entails direct DNA sampling of the catch. However, non-invasive monitoring is possible through analysis of so-called environmental DNA (abbreviated eDNA). Environmental DNA simply refers to genetic material, obtained directly from an environmental sample (water, air or soil), without any obvious parts of macro-organisms (Hansen et al. 2018). DNA is continuously shed by all organisms to the surrounding environment through skin (scales and mucus from fish), faeces, gametes, etc. (Hansen et al. 2018). Since marine organisms shed DNA “particles” directly into the sea, eDNA can be obtained from a water sample. The procedure is relatively simple: following water collection (normally ranging from typically 250 mL–5 L (Goldberg et al. 2016)), the water is filtered through a filter that retains the minute particles containing DNA, which can be extracted and analysed by either qPCR, ddPCR or meta-barcoding methods (Fig. 20.3). Quantitative PCR has been the preferred method to analyse the presence (and abundance) of specific species of interest (e.g., Thomsen et al. 2012b). For example, qPCR has been used extensively as a tool for the detection of rare and invasive species, which otherwise can be difficult to detect by conventional methods (e.g. Thomsen et al. 2012b). On the other hand, meta-barcoding has been the favoured tool for identifying and describing entire marine biological communities within or across taxonomic groups (e.g. Stat et al. 2017; Thomsen et al. 2012a, 2016). For such purposes, meta-barcoding is considered superior to qPCR because a large number of species can be targeted at the same time. For example, a recent study by Stat et al. (2017) tested several ‘universal’ PCR barcoding assays, including one for fish that recovered 69 different taxa, of which 33 of them were resolved to species.
The application of eDNA has been shown to be a cost-effective and sensitive sampling method (Sigsgaard et al. 2015) and shows comparable or even more accurate estimates of species diversity compared to established monitoring approaches like net fishing and diving (Thomsen et al. 2012a). Moreover, several studies have found positive correlations between species-specific DNA concentrations and biomass and abundance in controlled experimental settings like aquaria and ponds (Doi et al. 2015; Klymus et al. 2015), thus opening up possibilities for interesting applications. For example, it has been suggested that eDNA may be used as a future substitute or supplement to established stock assessments of commercial fish (Thomsen et al. 2016). Although we are still quite far from this scenario (Hansen et al. 2018), eDNA may be used in the future to reduce potential bycatch by focusing fishing efforts in areas with low numbers of potential bycatch species. In fact, Thomsen et al. (2016) have already tested such an application indirectly. They used a meta-barcoding approach to analyse seawater samples collected in Southwestern Greenland and compared it to trawling catch data. They generally observed weak but significant correlations between numbers of sequences belonging to specific groups of fish and biomass/abundance data, indicating a potential application for using eDNA as a tool for targeted fishing.
5.2 Current Limitations
Several issues exist which needs to be properly addressed before eDNA can be implemented to assist in more selective fishing efforts. First, faster ways of processing and analysing samples are needed to allow analysis directly at the fishing grounds. Such techniques, however, are already under development (see below section on future perspectives). Other more fundamental issues relate to (i) retention time of DNA (i.e. how long can it be detected in the environment) and (ii) its correlation with biomass. The first issue depends on the effects of abiotic and biotic factors that influence the production, degradation and transport of eDNA (see Hansen et al. (2018)). Here, abiotic factors like water temperature, pH and UV light determine the rate of DNA decay in the water, while biotic factors like size, age, species, health, sex and food condition influence eDNA production. Moreover, ocean currents will determine the distribution of eDNA, which may be carried hundreds of kilometres away from the source within days (Thomsen et al. 2012a). Still, marine eDNA studies report good concordance between eDNA analyses and visual surveys of local vertebrate communities (Port et al. 2016; Thomsen et al. 2012a). Nonetheless, environmental conditions play a large role in the production and turnover of eDNA and will likely vary significantly due to the widely different geospatial environmental conditions in our oceans (Klymus et al. 2015; Strickler et al. 2014).
An improved understanding of DNA production is crucial for assessing the relationship between the number of observed eDNA copies and the biomass in natural environments. While experimental studies in general have shown solid positive correlations between DNA and biomass (Doi et al. 2015; Klymus et al. 2015), relationships from natural systems are less pronounced, or in some cases even lacking (Spear et al. 2015; Thomsen et al. 2016). This is likely due to the complexity of such systems. For example, compared to a controlled experiment, natural habitats comprise many different size and age groups of fish, which will shed different amounts of DNA per mass, e.g. due to differences in metabolism, life stage or condition (Hansen et al. 2018). Moreover, different species of fish may shed more DNA than others, rendering it difficult to directly compare eDNA levels across species with widely different biology. Hence, before eDNA can be used in targeted fisheries, more empirical research is needed. This will improve our understanding about how these issues affect eDNA quantification and pave the way for future research into fisheries application.
5.3 Case-Study
In a current study, conducted by DTU Aqua, Denmark, the potential for using eDNA to avoid bycatch is being directly investigated. Specifically, this study aims at testing the potential for applying qPCR as a tool for monitoring absolute and relative biomass of European sprat (Sprattus sprattus), Atlantic herring (Clupea harengus) and sand eel (Ammodytidae). While all species are harvested commercially, the bycatch of herring is a well-known problem, especially in sprat fisheries, and to some extent, in sand eel fisheries. Currently, bycatch of herring are discarded when the bycatch quota is exceeded. However, with the introduction of the Landing Obligation, all bycatch of herring should, in theory, be deducted from this quota. This may lead to a scenario where herring is a “choke species” for sprat and sand eel fisheries leading to their closure. Thus, approaches facilitating more selective fisheries are warranted by the industry.
In the study, water samples were collected on board the fishing vessels while fishing and estimates of catch and bycatch were conducted. Subsequently the water samples were analysed for the quantity of species-specific DNA to assess potential correlations. The data are still being analysed, thus only a few preliminary results are available. However, they demonstrate large differences in the relative proportion of DNA from Atlantic herring in the European sprat fisheries across fishing localities (Fig. 20.5). Whether this is also well correlated with the actual biomass caught is currently being investigated.
6 Future Perspectives
Some of the major limitations of the methods described in this chapter are their accessibility, time from sampling to results, the required manual labour and associated cost. Most of the techniques are currently only performed by highly trained personnel in well-equipped molecular laboratories. However, in the near future, it is possible that people with little molecular training can use many of the techniques in situ, e.g. on board fishing vessels or at fishing ports. This should lower the costs and facilitate faster communication between fishers and managers regarding bycatch and catch composition, potentially leading to more efficient fisheries management and conservation. On board analyses of eDNA in water samples prior to fishing would, moreover, enhance the potential of using eDNA for targeted fishing efforts, as it would allow near real-time analysis at the fishing grounds.
Already today, novel portable devices and kits exist that allow for DNA extraction and sequencing outside the laboratory. For example, Oxford Nanopore Technologies has designed a device for automated sample preparation and even PCR (https://nanoporetech.com/products/voltrax). Combined with small portable sequencing machines like, for example, the MinION or the smartphone compatible SmidgION (Oxford Nanopore Technologies), this allows for fast onsite DNA barcoding (Menegon et al. 2017). The MinION device has the same size as a normal smart phone but can sequence up to 20 Gigabases of DNA (20 billion A, C, T and G’s) per run. The sequences are directly uploaded to a laptop computer and can therefore be analysed in close-to-real-time. The major disadvantage of the MinION is a high sequencing error rate, compared to laboratory DNA sequencing. However, this is to some degree neutralized by the large sequencing output and recent development in analytical methods. This poses another challenge, as analysis of genomic data is computationally intensive and hence some applications may need internet access to upload the data to an online server for fast on site analysis (Srivathsan et al. 2018) and subsequent storing of results. Moreover, automated methods and software, that can transfer gigabytes of DNA data from these devices into a format that can be interpreted by the non-expert (fishers), is a field, which needs more research and development. Further, the cost per run is still high compared to conventional DNA barcoding, however; sequencing methods exist for tagging, pooling and subsequent sequencing of up to hundreds of samples per MinION run, thereby lowering the cost per sample significantly (Srivathsan et al. 2018). Another option is stopping the sequencing run when just enough sequences have been obtained for producing proper species identification. Subsequently, the device can be washed and used again until reaching the overarching sequencing capacity.
Onsite analysis of species composition and quantification in mixed species products or eDNA may also be conducted using qPCR. For example, the US-based company Biomeme (https://biomeme.com) has developed an ultra-small qPCR device that attaches to a smartphone and can run up to nine samples with up to two different target species within an hour (e.g. sprat and herring). The same company has also developed fast DNA extraction kits that allows for DNA preparation within a few minutes without need of any lab equipment. This kit uses disposable, single-use consumables, which decreases the possibility for DNA contamination among samples. Lastly, future eDNA analysis may also be conducted by so called “ecogenomic sensors”, which are submersible instruments for autonomous in situ automatic DNA analysis of water samples (Scholin 2009). The 2nd generation environmental sample processor (2G ESP) is such a device and is essentially a DNA lab in a waterproof can. The 2G ESP has already shown practical applications for monitoring microorganisms (Ottensen et al. 2014), zooplankton (Harvey et al. 2012) and eDNA from fish (personal communication, Brian K. Hansen)). The 2G ESP can be moored to a specific location (e.g. a dockside or buoy), be mobile (free drifting) or carried on board fishing vessels where it can conduct automatized water sampling, DNA extraction and qPCR analysis. The ESP provides two-way communications, which allows the end-user to remotely assess the results in near real-time. This has potential interesting applications for fisheries, as this may allow for eDNA analysis directly at the fishing grounds, which then can be communicated to the fishing fleet and act as a supplement in order to avoid bycatch.
In conclusion, DNA-based methods can already provide important information that can be used under the Landing Obligation to identify and prevent bycatch as well as in control, management and consumer protection. The rapid and continuous development of portable and automatic real-time devices for DNA analysis are expected to become more powerful, user-friendly and much cheaper in the near future. This will likely increase the incentives to use genetics in fisheries management, including subjects related to discards.
References
April, J., Mayden, R.L., Hanner, R.H., Bernatchez, L. (2011). Genetic calibration of species diversity among North America’s freshwater fishes. Proceedings of the National Academy of Sciences of the United States of America, 108, 10602–10607. https://doi.org/10.1073/pnas.1016437108.
Arruda, L.F., Borghesi, R., Oetterer, M. (2007). Use of fish waste as silage: A review. Brazilian Archives of Biology and Technology, 50(5), 879–886. https://doi.org/10.1590/S1516-89132007000500016.
Ballard, J.W.O., & Whitlock M.C. (2004). The incomplete natural history of mitochondria. Molecular Ecology, 13, 729–744. https://doi.org/10.1046/j.1365-294X.2003.02063.x.
Ballin, N.Z., Vogensen, F.K., Karlsson A.H. (2009). Species determination – Can we detect and quantify meat adulteration? Meat Science, 83, 165–174. https://doi.org/10.1016/j.meatsci.2009.06.003.
Bojolly, D., Doyen, P., Le Fur, B., Christaki, U., Verrez-Bagnis, V., Gard, T. (2017). Development of a qPCR method for the identification and quantification of two closely related tuna species, bigeye tuna (Thunnus obesus) and yellowfin tuna (Thunnus albacares), in canned tuna. Journal of Agricultural and Food Chemistry, 65, 913–920. https://doi.org/10.1021/acs.jafc.6b04713.
Burg, T.M. (2007). Genetic analysis of wandering albatrosses killed in longline fisheries off the east coast of New Zealand. Aquatic Conservation-Marine and Freshwater Ecosystems, 17, 93–101. https://doi.org/10.1002/aqc.907.
Cole, L.W. (2016). The evolution of per-cell organelle number. Frontiers in Cell and Development, 4, 85. https://doi.org/10.3389/fcell.2016.00085.
Coyne, J., & Orr, H. (2004). Speciation. Sunderland: Sinnauer Associates.
Doi, H., Uchii, K., Takahara, T., Matcuhashi, S., Yamanaka, H., Minamoto, T. (2015). Use of droplet digital PCR for estimation of fish abundance and biomass in environmental DNA surveys. PLoS One, 10. https://doi.org/10.1371/journal.pone.0122763.
Floren, C., Wiedemann, I., Brenig, B., Schutz, E., & Beck, J. (2015). Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food Chemistry, 173, 1054–1058. https://doi.org/10.1016/j.foodchem.2014.10.138.
Galtier, N., Nabholz, B., Glemin, S., Hurst, G.D.D. (2009). Mitochondrial DNA as a marker of molecular diversity: A reappraisal. Molecular Ecology, 18, 4541–4550. https://doi.org/10.1111/j.1365-294X.2009.04380.x.
Goldberg, C.S., Turner, C.R., Deiner, K., Klymus, K.E., Thomsen, P.F., Murphy, M.A., et al. (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods in Ecology and Evolution, 7, 1299–1307. https://doi.org/10.1111/2041-210X.12595.
Goodwin, S., McPherson, J.D., McCombie, W.R. (2016). Coming of age: Ten years of next-generation sequencing technologies. Nature Review Genetics, 17, 333–351. https://doi.org/10.1038/nrg.2016.49.
Hansen, B.H., Bekkevold, D., Clausen, L.W., & Nielsen, E.E. (2018). The sceptical optimist: Challenges and perspectives for the application of environmental DNA in marine fishes. Fish and Fisheries, 1, 1–18. https://doi.org/10.1111/faf.12286.
Harvey, J.B.J., Ryan, J.P., Marin, R., Preston, C.M., Alvarado, N., Scholin, C.A., et al. (2012). Robotic sampling, in situ monitoring and molecular detection of marine zooplankton. Journal of Esperimental Marine Biology and Ecology, 413, 60–70. https://doi.org/10.1016/j.jembe.2011.11.022.
Hebert, P.D.N., Ratnasingham, S., deWaard, J.R. (2003). Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society B-Biological Sciences, 270, S96−S99. https://doi.org/10.1098/rsbl.2003.0025.
Helyar, S.J., Lloyd, H.A., de Bruyn, M., Leake, J., Bennett, N., Carcalho, G.R. (2014). Fish product mislabelling: Failings of traceability in the production chain and implications for illegal, unreported and unregulated (IUU) fishing. PLoS One, 9, https://doi.org/10.1371/journal.pone.0098691.
Holden, M.J. (1973). Are long-term sustainable fisheries for elasmobranchs possible? Rapports et Procés Verbaux des Rèunionsdu Conseil International pour l’Exploration de la Mer, 164, 360–367.
Hopkins, G.W., & Freckleton, R.P. (2002). Declines in the numbers of amateur and professional taxonomists: Implications for conservation. Animal Conservation, 5, 245–249. https://doi.org/10.1017/S1367943002002299.
Huxley-Jones, E., Shaw, J.L.A., Fletcher, C., Parnell, J., Watts, P.C. (2012). Use of DNA barcoding to reveal species composition of convenience seafood. Conservation Biology, 26, 367–371. https://doi.org/10.1111/j.1523-1739.2011.01813.x.
Iñarra, B., Bald, C., Cebrián, M., Antelo, L.T., Franco-Uría, A., Vázquez, J.A., et al. (this volume). What to do with unwanted catches: Valorisation options and selection strategies. In S.S. Uhlmann, C. Ulrich, S.J. Kennelly (Eds.), The European Landing Obligation – Reducing discards in complex, multi-species multi-jurisdictional fisheries. Cham: Springer.
Ivanova, N.V., Zemlak, T.S., Hanner, R.H., Hebert, P.D.N. (2007). Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes, 7, 544–548. https://doi.org/10.1111/j.1471-8286.2007.01748.x.
Kelleher, K. (2005). Discards in the world’s marine fisheries. An update. FAO Fisheries Technical Paper. No. 470. Rome, FAO. Available at: http://www.fao.org/3/a-y5936e.pdf
Klymus, K.E., Richter, C.A., Chapman, D.C., Paukert, C. (2015). Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biological Conservation, 183, 77–84. https://doi.org/10.1016/j.biocon.2014.11.020.
Larsen, E., Dalskov, J., Nielsen, E.E., Kirkegaard, E., Nielsen, J.W., Tørring, P., et al. (2013). Dansk fiskeris udnyttelse af discardforbuddet: En udredning. Charlottenlund: DTU Aqua. Institut for Akvatiske Ressourcer. DTU Aqua-rapport; No. 275-2013, (p. 106).
Lockley, A.K., & Bardsley, R.G. (2000). DNA-based methods for food authentication. Trends in Food Science & Technology, 11, 67−77. https://doi.org/10.1016/S0924-2244(00)00049-2.
Mayr, E. (1942). Systematics and the origin of species. New York: Columbia University Press.
Mayr, E. (1963). Animal species and evolution. Cambridge, MA: Harvard University Press.
McEachran, J.D., & Musick, J.A. (1973). Characters for distinguishing between immature specimens of the sibling species, Raja erinacea and Raja ocellata (Pisces: Rajidae). Copeia, 238–250. https://doi.org/10.2307/1442962.
Menegon, M., Cantaloni, C., Rodriguez-Prieto, A., Centomo, C., Abdelfattah, A., Rossato, M., et al. (2017). On site DNA barcoding by nanopore sequencing. PLoS One, 12, https://doi.org/10.1371/journal.pone.0184741.
Murray, D.C., Bunce, M., Cannell, B.L., Oliver, R., Houston, J., White, N.E., et al. (2011). DNA-based faecal dietary analysis: A comparison of qPCR and high throughput sequencing approaches. PLoS One, 6, https://doi.org/10.1371/journal.pone.0025776.
Nielsen, E.E. (2016) Population or point-of-origin identification. In A.M. Naarum & R.H. Hanner (Eds.), Seafood authenticity and traceability (pp. 149–169). London: Academic.
Ottensen, E.A., Young, C.R., Gifford, S.M. Eppley, J.M., Marin, R., Schuster, S.C., et al. (2014). Multispecies diel transcriptional oscilliations in open oceab heterotrophic bacterial assemblages. Science, 345, 207–212. https://doi.org/10.1126/science.1252476.
Ovenden, J.R., Berry, O., Welch, D.J., Buckworth, R.C., Dichmont, C.M. (2015). Ocean’s eleven: A critical evaluation of the role of population, evolutionary and molecular genetics in the management of wild fisheries. Fish and Fisheries, 16, 125–159. https://doi.org/10.1111/faf.12052.
Pepe, T., Trotta, M., Di Marco, I., Anastasio, A., Bautista, J.M., Cortesi, M.L. (2007). Species identification in surimi-based products. Journal of Agricultural and Food Chemistry, 55, 3681−3685. https://doi.org/10.1021/jf063321o.
Pompanon, F., Deagle, B.E., Symondson, W.O.C., Brown, D.S., Jarman, S.N., Tarberlet, P. (2012). Who is eating what: Diet assessment using next generation sequencing. Molecular Ecology, 21, 1931–1950. https://doi.org/10.1111/j.1365-294X.2011.05403.x.
Port, J.A., O’Donnell, J.L., Romero-Maraccini, O.C., Leary, P.R., Litvin, S.Y., Nickols, K.H., et al. (2016). Assessing vertebrate biodiversity in a kelp forest ecosystem using environmental DNA. Molecular Ecology, 25, 527–541. https://doi.org/10.1111/mec.13481.
Ratnasingham, S., & Hebert, P.D.N. (2007). BOLD: The barcode of life data system (http://www.barcodinglife.org). Molecular Ecology Notes, 7, 355–364. https://doi.org/10.1111/j.1471-8286.2007.01678.x.
Saraste, M. (1999). Oxidative phosphorylation at the fin de siecle. Science, 283, 1488−1493. https://doi.org/10.1126/science.283.5407.1488.
Scholin, C.A. (2009). What are “ecogenomic sensors?”—A review and thoughts for the future. Ocean Science Discussions, 6, 191–213. https://doi.org/10.5194/os-6-51-2010.
Scientific, Technical and Economic Committee for Fisheries (STECF). (2017). Long-term management of skates and rays (STECF-17-21). Publications Office of the European Union, Luxembourg, 2017, ISBN 978-92-79-67493-8. https://doi.org/10.2760/44133, JRC109366.
Sigsgaard, E.E., Carl, H., Møller, P.R., & Thomsen, P.F. (2015). Monitoring the near-extinct European weather loach in Denmark based on environmental DNA from water samples. Biological Conservation, 183, 46–52. https://doi.org/10.1016/j.biocon.2014.11.023.
Spear, S.F., Groves, J.D., Williams, LA., Waits, L.P. (2015). Using environmental DNA methods to improve detectability in a hellbender (Cryptobranchus alleganiensis) monitoring program. Biological Conservation, 183, 38–45. https://doi.org/10.1016/j.biocon.2014.11.016.
Srivathsan, A., Baloğlu, B., Wang, W., Tan, W.Z., Bertrand, D., Ng, A.H.G., et al. (2018). A MinION™-based pipeline for fast and cost-effective DNA barcoding. Molecular Ecology Resources. https://doi.org/10.1111/1755-0998.12890.
Stat, M., Huggett, M.J., Bernasconi, R., DiBattista, J.D. Berry, T.E., Newman S.J., et al. (2017). Ecosystem biomonitoring with eDNA: Metabarcoding across the tree of life in a tropical marine environment. Scientific Reports, 7. https://doi.org/10.1038/s41598-017-12501-5.
Strickler, K.M., Fremier, A.K., Goldberg, C.S. (2014). Quantifying effects of UV- B, temperature, and pH on eDNA degradation in aquatic microcosms. Biological Conservation, 183, 85–92. https://doi.org/10.1016/j.biocon.2014.11.038.
Teletchea, F. (2009). Molecular identification methods of fish species: Reassessment and possible applications. Reviews in Fish Biology and Fisheries, 19, 265–293. https://doi.org/10.1007/s11160-009-9107-4.
Thomas, A.C., Deagle, B.E., Eveson, J.P., Harsch, C.H., & Trites, A.W. (2016). Quantitative DNA metabarcoding: Improved estimates of species proportional biomass using correction factors derived from control material. Molecular Ecology Resources, 16, 714–726. https://doi.org/10.1111/1755-0998.12490.
Thomsen, P.F., Kielgast, J., Iversen, L.L., Møller, P.R., Rasmussen, M., Willerslev, E. (2012a). Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS One, 7, https://doi.org/10.1371/journal.pone.0041732.
Thomsen, P.F., Kielgast, J., Iversen, L.L., Wiuf, C., Rasmussen, M., Gilbert, M.T., et al. (2012b). Monitoring endangered freshwater biodiversity using environmental DNA. Molecular Ecology, 21, 2565–2573. https://doi.org/10.1111/j.1365-294X.2011.05418.x.
Thomsen, P.F., Møller, P.R., Sigsgaard, E.E., Knudsen, S.W., Jørgensen, O.A., Willerslev, E. (2016). Environmental DNA from seawater samples correlate with trawl catches of subarctic, deepwater fishes. PLoS One, 11. https://doi.org/10.1371/journal.pone.0165252.
Viðarsson, J.R., Larsen, E.P., Valeiras, J., & Ragnarsson, S.Ö. (this volume). Onboard and vessel layout modifications. In S.S. Uhlmann, C. Ulrich, S.J. Kennelly (Eds.), The European Landing Obligation – Reducing discards in complex, multi-species and multi-jurisdictional fisheries. Cham: Springer.
Walsh, H.E., & Edwards, S.V. (2005). Conservation genetics and Pacific fisheries by-catch: Mitochondrial differentiation and population assignment in black-footed albatrosses (Phoebastria nigripes). Conservation Genetics, 6, 289–295. https://doi.org/10.1007/s10592-004-7824-8.
Ward, R.D., Zemlak, T.S., Innes, B.H., Last, P.R., Hebert, P.D.N. (2005). DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B-Biological Sciences, 360, 1847–1857. https://doi.org/10.1098/rstb.2005.1716.
Wheeler, Q.D., Raven, P.H., Wilson, E.O. (2004). Taxonomy: Impediment or expedient? Science, 303, 285–285. https://doi.org/10.1126/science.303.5656.285.
Acknowledgments
This work has received funding from the Horizon 2020 Programme under grant agreement DiscardLess number 633680. This support is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2019 The Author(s)
About this chapter
Cite this chapter
Jacobsen, M.W., Hansen, B.K., Nielsen, E.E. (2019). Possible Uses of Genetic Methods in Fisheries Under the EU Landing Obligation. In: Uhlmann, S., Ulrich, C., Kennelly, S. (eds) The European Landing Obligation. Springer, Cham. https://doi.org/10.1007/978-3-030-03308-8_20
Download citation
DOI: https://doi.org/10.1007/978-3-030-03308-8_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-03307-1
Online ISBN: 978-3-030-03308-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)