Abstract
Segmentation of the left atrium and deriving its size can help to predict and detect various cardiovascular conditions. Automation of this process in 3D Ultrasound image data is desirable, since manual delineations are time-consuming, challenging and observer-dependent. Convolutional neural networks have made improvements in computer vision and in medical image analysis. They have successfully been applied to segmentation tasks and were extended to work on volumetric data. In this paper we introduce a combined deep-learning based approach on volumetric segmentation in Ultrasound acquisitions with incorporation of prior knowledge about left atrial shape and imaging device. The results show, that including a shape prior helps the domain adaptation and the accuracy of segmentation is further increased with adversarial learning.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Quantification of cardiac chambers and their functions stay the most important objective of cardiac imaging [7]. Left atrium (LA) physiology and function have an impact on the whole heart performance and its size is a valuable indicator for various cardiovascular conditions, such as atrial fibrillation (AF), stroke and diastolic dysfunction [7]. Compared to cardiac computed tomography (CCT) and cardiac magnetic resonance (CMR), as modalities to examine the heart, echocardiography provides wide availability, safety and good spatial and temporal resolution, without exposing the patients to harmful radiation. Volumetric measurements consider changes in all spatial dimensions, however, to obtain reproducible and accurate three-dimensional (3D) measurements, requires expert experience and is time consuming [4]. Automated segmentation and quantification could help to reduce inter/intra-observer variabilities and might also save costs and time in echocardiographic laboratories [4].
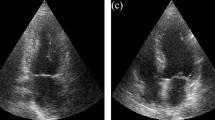
Row 1: ground truth segmentation of LA, Row 2: prediction by DGA architecture (Table 3), Column 1 & 2: device EPIQ 7C (trained on Vivid E9), Column 3 & 4: device Vivid E9 (trained on EPIQ 7C), Column 5: device iE33 (trained on Vivid E9).
Previous automatic and semi-automatic approaches for LA segmentation have focused CCT and CMR as a planning and guidance tool for LA catheter interventions [1]. For 3D Ultrasound (US), the left ventricle (LV) was the segmentation target, since its size and function remain the most important indication for a cardiac study [6]. LA segmentation in 3D US has not received much attention, apart from commercially available methods, which were also successfully validated against the gold standard CMR and CCT [3, 10]. Almeida et al. [1] adapted a segmentation framework for LV, based on B-spline explicit active surfaces. Those methods, however, require more or less manual interaction. Recently, fully automatic segmentation of the left heart was validated against 2D and 3D echocardiography, as well as CCT [4].
Convolutional neural networks (CNN) and their special architectures of fully convolutional networks (FCN) have successfully been applied to the problem of medical image segmentation. Those networks are trained end-to-end, process the whole image and perform pixel-wise classification. The V-Net extends this idea to volumetric image data and enables 3D segmentation with the help of spatial convolutions, instead of processing the volumes slice-wise [8].
Automated segmentation in cardiac US images is challenging, due to artifacts caused by respiratory motion, shadows or signal-dropouts. Including shape priors in this task can help algorithms to yield more accurate and anatomically plausible results. Oktay et al. [9] introduced a way to incorporate such a prior with the help of an autoencoder network, that leads segmentation masks to follow an underlying shape representation.
Image data might be different (e.g with respect to resolution, contrast), due to varying imaging protocols and device manufacturers [2, 5]. Although the segmentation task is equivalent, neural networks perform poorly when applied to data that was not available during training. Generating ground truth maps and retraining a new model for each domain is not a scalable solution. The problem of models to generalize to new image data can be approached by domain adaptation. Kamnitsas et al. [5] successfully introduced the application of unsupervised domain adaptation for different MRI databases, when an adversarial neural network was influencing the feature maps of a CNN, which was employed for a segmentation task.
In this work, LA segmentation in 3D US volumes is performed with the help of neural networks. For the volumetric segmentation, V-Net will be trained, combined with additional losses, taking into account the geometrical constraint introduced by the shape of the LA and the desired ability to generalize to different US devices and settings.
2 Methodology
Our framework, as depicted in Fig. 2, consists of three existing methods; 3D Fully Convolutional Segmentation Network [8], Anatomic Constraint [9], and Domain Adaptation [5]. Nevertheless, it is a novelty to model the solution in a single framework, enabling analysis on the contribution of each element on the primary segmentation task. Further, the domain adaptation method has been leveraged to a 3D FCN segmentation framework, and applied successfully to the LA, showing a statistical significant improvement, as reported in Sect. 3.
Overview of the combined architecture: Image data \(X_i\) is processed by V-Net [8]. \(\mathcal {L}_{seg}\) is calculated from the resulting segmentation \(\hat{Y}_i\) and the ground truth \(Y_i\). Additionally, \(\hat{Y}_i\) and \(Y_i\) are encoded (E) to get the shape constraint. An optional number of feature maps, based on \(X_i\) are extracted from V-Net to be processed in the classifier (C), which predicts a domain \(\hat{d}_i\). Cross-entropy between \(\hat{d}_i\) and the real domain \(d_i\) determines the adversarial loss.
Segmentation. For the segmentation task, we employ V-Net [8] as a 3D FCN, which processes an image volume of size n, \(X_i = \{x_1,...,x_n\}\), \(x_i \in \mathcal {X}\) and yields a segmentation mask \(\hat{Y}_i = \{\hat{y}_1,...,\hat{y}_n\}\), \(\hat{y}_i \in \mathcal {\hat{Y}}\) in the original resolution. \(\mathcal {X}\) represents the feature space of US acquisitions and \(\mathcal {\hat{Y}}\) describes the probability of a voxel belonging to the segmentation.
The objective function of V-Net is adapted to the segmentation task. It is based on the Dice coefficient (Eq. 1), taking into account the possible imbalance of foreground to background, alleviating the need to re-weight samples.
with \(\hat{y}_i\) being the prediction and \(y_i\) the voxels of the ground truth \(Y_i\) from the binary distribution \(\mathcal {Y}\).
Shape Prior. Incorporation of the shape prior to help the segmentation task is realized by training an autoencoder network on the segmentation ground truth masks Y. The encoder reduces the label to a latent, low resolution representation \(E(Y_i)\) and the decoder tries to retrieve the original volume \(Y_i\). Due to the resolution reduction of the encoder, the shape information is encoded in a compact fashion [9].
During training, the output of the segmentation network \(\hat{Y}_i\) is passed to the encoder, along with the ground truth label \(Y_i\). Based on a distance metric d(\(\cdot \),\(\cdot \)), a loss between the latent codes of both inputs is calculated as
The gradient is then back-propagated to the segmentation network.
Domain Adaptation. When a network is trained on one type of data \(\mathcal {X}_S\) (source domain) and evaluated on another \(\mathcal {X}_T\) (target domain), the performance is poor in most cases. Domain invariant features are desired to make the segmentation network perform well on different data sets. Kamnitsas et al. [5] propose an approach to generate domain invariant features to increase a networks generalization capability.
Processing an image volume in a CNN yields a latent representation \(h_l(X_i)\) after convolutional layer l. If the network is not domain invariant, those feature maps contain information about the data type (source or target domain). The idea to solve this issue, is to train a classifier C, which takes feature maps of the segmentation network as input and returns whether the input data was from source (\(X_S\)) or target (\(X_T\)) domain: \(C(h_l(X_i)) = \hat{d}_i \in \{S,T\}\). The accuracy of this classifier is an indicator of how domain invariant the features are.
Combination. The ideas introduced in the previous sections are combined to exploit the advantages of the individual approaches (Fig. 2). The loss of the domain classifier is used as an adversarial loss term, since the goal of the segmentation network is to lower the classification accuracy (i.e maximize its loss). The inability of the classifier to tell, which type of data was segmented means that the feature maps are domain invariant. At the same time, \(\mathcal {L}_{seg}\) and \(\mathcal {L}_{enc}\) should be minimized. With \(\mathcal {L}_{adv}\) as the binary cross entropy loss of the classifier C, this yields the following combined loss function:
3 Experiments and Results
To evaluate the influence of different loss terms, we apply it to 3D Ultrasound data to perform end-systolic LA segmentation. The network is trained with images and labels from one device and tested on different devices.
Dataset. The data available for this work are 3D transthoracic echocardiography (TTE) examinations taken from clinical routine, which brings variations from differences in US imaging devices, protocols (resolution, opening angle) and patients (healthy, abnormal), raising the necessity of our proposed framework (Table 1). Multiple international centers contributed to a pool of 161 datasets, containing the LA ground truth segmentation in the entire recorded heart cycle, with the relevant phases for LA functionality (end-diastole, end-systole and pre-atrial contraction) identified.
Acquisition was performed with systems from GE (Vivid E9, GE Vingmed Ultrasound) and Philips (EPIQ 7C and iE33, Philips Medical Systems), each equipped with a matrix array transducer. Since there are only 15 datasets for device iE33, those examinations are not used for training, only for evaluation. The data is down-sampled, preserving angles and ratios, by zero padding (cf. Fig. 1), to enable processing of the entire volumes.
Implementation. Network architectures are implemented using the TensorFlowFootnote 1 library (version 1.4) with GPU support. For our approach, the V-Net architecture is adapted, such that volumes of size \(64\,\times \,64\,\times \,64\) can be processed. The autoencoder network architecture is inspired from the one proposed in [9]. Feature maps from different levels and sizes are extracted from V-Net to be processed in the classifier (Fig. 2). By (repeated) application of convolutions of filter size 2 with stride 2, the feature maps are brought to the V-Net valley size (\(4\,\times \,4\,\times \,4\)), so they can be concatenated along the channel dimension.
Training Details. The autoencoder network is trained before the combined training procedure, to obtain a meaningful latent representation for the shape prior. In the following training stages, the parameters of this network are frozen. The segmentation network is shortly pre-trained, as well as the classifier to introduce stability in the combined training and it can focus on realizing the scenario defined by the settings of \(\lambda _{enc}\) and \(\lambda _{adv}\). Feature maps L0, L2, M, R2 and R0 of the segmentation network are extracted for the classifier.
The combined training procedure starts by adding \(\mathcal {L}_{enc}\), for incorporation of the shape prior to the segmentation loss \(\mathcal {L}_{seg}\). Adversarial influence begins after 10 epochs of combined training, linearly increasing \(\lambda _{adv}\) until it reaches its maximum of 0.001 after another 10 epochs. While the combined training exclusively adjusts the parameters of the segmentation network \(\theta _{seg}\), the classifier parameters \(\theta _{adv}\) are continued to be trained in parallel to retain a potent adversarial loss term. A training overview is given in Table 2.
Evaluation. The segmentation network returns a volume \(\hat{Y}_i\) of probabilities for the voxels to belong to the foreground, i.e the segmentation of the LA. The threshold for the cutoff probability to obtain a binary segmentation mask is determined by the best Dice coefficient on the validation set, from which the biggest connected component is selected as the final LA segmentation.
Segmentation metrics [1, 9] are reported in Table 3 for the recommended phase of LA segmentation (end-systole ES [7]). We refer to the V-Net architecture with the additional loss term \(\mathcal {L}_{enc}\), calculated from the L2-distance (\(d(p,q) = \Vert p - q\Vert ^2_2\)), as geometry agnostic CNN GAL2. To investigate the influence of a different distance metric, GAACD uses the angular cosine distance, as it was proposed in [2] (ACD, \(d(p,q) = 1 - \frac{\sum _{i}p_i \cdot q_i}{\Vert p\Vert _2 \cdot \Vert q\Vert _2}\)). Our domain and geometry agnostic CNN DGA leverages the better performing distance metric (ACD, based on test results) with the adversarial loss \(\mathcal {L}_{adv}\). We define statistical significance based on the paired two-sample t-test on a 5% significance level.
When training on EPIQ 7C, V-Net performs better than the other architectures on the same device. However, those margins are not statistically significant (MSD: \(p = 0.65\), HD: \(p = 0.24\), DC: \(P = 0.66\)), compared to DGA. The increased performance of DGA compared to V-Net and ACNN is significant with respect to all metrics. Vivid E9 training yields V-Net with the best performance on the same device, with statistical significance on all metrics. DGA is significantly outperforming V-Net on EPIQ 7C in terms of MSD and HD. No significant differences are observable on the evaluation of device iE33. Independent of the distance metric utilized, an improvement in generalizability is observable compared to V-Net when the shape prior is included (GAL2 & GAACD).
4 Discussion and Conclusion
While V-Net performs well on the task of LA segmentation, the ability to generalize to new domains is achieved by the introduction of a shape prior and the adversarial loss, as shown in the results. Including the shape prior boosts the segmentation performance on unseen devices and theoretically leads to a geometrically plausible segmentation in case of image artifacts. We ensure a potent classifier by training it in parallel to the DGA architecture. Thus, it can detect domain-specific features throughout the training procedure. The distance metric for the geometrical constraint is an interesting subject to further investigate, as well as extracting different V-Net-layers for processing in the classifier network.
Notes
References
Almeida, N., et al.: Left-atrial segmentation from 3-D ultrasound using B-spline explicit active surfaces with scale uncoupling. IEEE UFFC-S 63(2), 212–221 (2016)
Baur, C., Albarqouni, S., Navab, N.: Semi-supervised deep learning for fully convolutional networks. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 311–319. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_36
Buechel, R.R., et al.: Assessment of left atrial functional parameters using a novel dedicated analysis tool for real-time three-dimensional echocardiography: validation in comparison to magnetic resonance imaging. Int. J. Cardiovas Imag. 29(3), 601–608 (2013)
van den Hoven, A.T., et al.: Transthoracic 3D echocardiographic left heart chamber quantification in patients with bicuspid aortic valve disease. Int. J. Cardiovas Imag. 33(12), 1895–1903 (2017)
Kamnitsas, K., et al.: Unsupervised domain adaptation in brain lesion segmentation with adversarial networks. In: Niethammer, M., et al. (eds.) IPMI 2017. LNCS, vol. 10265, pp. 597–609. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59050-9_47
Knackstedt, C., et al.: Fully automated versus standard tracking of left ventricular ejection fraction and longitudinal strain: the FAST-EFs multicenter study. JACC 66(13), 1456–1466 (2015)
Lang, R.M., et al.: Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the american society of echocardiography and the european association of cardiovascular imaging. Eur. Heart J. 16(3), 233–271 (2015)
Milletari, F., Navab, N., Ahmadi, S.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571 (2016)
Oktay, O., et al.: Anatomically constrained neural networks (ACNNs): application to cardiac image enhancement and segmentation. IEEE TMI 37(2), 384–395 (2018)
Rohner, A., et al.: Functional assessment of the left atrium by real-time three-dimensional echocardiography using a novel dedicated analysis tool: initial validation studies in comparison with computed tomography. Eur. J. Echocardiogr. 12(7), 497–505 (2011)
Acknowledgment
We would like to thank Ozan Oktay for sharing his implementation of the ACNN architecture as a baseline of our project. Further, we thank Georg Schummers and Matthias Friedrichs from TOMTEC Imaging Systems GmbH for their support and helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Degel, M.A., Navab, N., Albarqouni, S. (2018). Domain and Geometry Agnostic CNNs for Left Atrium Segmentation in 3D Ultrasound. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11073. Springer, Cham. https://doi.org/10.1007/978-3-030-00937-3_72
Download citation
DOI: https://doi.org/10.1007/978-3-030-00937-3_72
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00936-6
Online ISBN: 978-3-030-00937-3
eBook Packages: Computer ScienceComputer Science (R0)