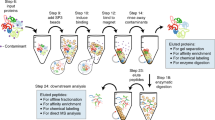
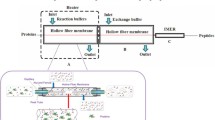
Abstract
The success of a proteomics experiment critically depends on the correct execution of the sample preparation. Gel-based sample preparation is a well-proven method widely used in qualitative and quantitative proteomics. Recently, sample preparation in a single vessel has gained popularity because of the ease to perform, the possibility for large-scale analysis, and the potential to improve sensitivity. In this chapter I describe two sample preparation protocols, filter-aided sample preparation (FASP) and single-pot solid-phase sample preparation (SP3).
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chen N, Pandya NJ, Koopmans F, Castelo-Szekelv V, van der Schors RC, Smit AB, Li KW (2014) Interaction proteomics reveals brain region-specific AMPA receptor complexes. J Proteome Res 13:5695–5706
Sielaff M, Kuharev J, Bohn T, Hahlbrock J, Bopp T, Tenzer S, Distler U (2017) Evaluation of FASP, SP3, and iST protocols for proteomic sample preparation in the low microgram range. J Proteome Res 16:4060–4072
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362
Hughes CS, Foehr S, Garfield DA, Furlong EE, Steinmetz LM, Krijgsveld J (2014) Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol 10:757
Moggridge S, Sorensen PH, Morin GB, Hughes CS (2018) Extending the compatibility of the SP3 paramagnetic bead processing approach for proteomics. J Proteome Res 17:1730–1740
Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11:319–324
Taoka M, Fujii M, Tsuchiya M, Uekita T, Ichimura T (2017) A sensitive microbead-based organic media-assisted method for proteomics sample preparation from dilute and denaturing solutions. ACS Appl Mater Interfaces 9:42661–42667
Ludwig KR, Schroll MM, Hummon AB (2018) Comparison of in-solution, FASP, and S-trap based digestion methods for bottom-up proteomic studies. J Proteome Res 17(7):2480–2490
Zougman A, Selby PJ, Banks RE (2014) Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics 14:1006–1000
Yu Y, Bekele S, Pieper R (2017) Quick 96FASP for high throughput quantitative proteome analysis. J Proteome 166:1–7
Pandya NJ, Klaassen RV, van der Schors RC, Slotman JA, Houtsmuller A, Smit AB, Li KW (2016) Group 1 metabotropic glutamate receptors 1 and 5 form a protein complex in mouse hippocampus and cortex. Proteomics 16:2698–2705
Gonzalez-Lozano MA, Koopmans F, Paliukhovich I, Smit AB, Li KW (2019) A fast and economical sample preparation protocol for interaction proteomics analysis. Proteomics:e1900027
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Li, K.W. (2019). Sample Preparation for Proteomics Analysis: Filter-Aided Sample Preparation (FASP) and Single-Pot Solid-Phase Sample Preparation (SP3). In: Li, K. (eds) Neuroproteomics. Neuromethods, vol 146. Humana, New York, NY. https://doi.org/10.1007/978-1-4939-9662-9_6
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9662-9_6
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-4939-9661-2
Online ISBN: 978-1-4939-9662-9
eBook Packages: Springer Protocols