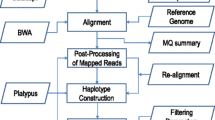
Abstract
Genotyping by sequencing (GBS) is a relatively new method used to determine the differences in the genetic makeup of individuals. Its novelty stems from a combination of two already available methods: genotyping and next-generation sequencing. Depending on the individual study design GBS protocols can take multiple forms, however most share a sequence of core steps that have to be undertaken. These include: sequencing of the DNA from the individuals of interest (usually two parents of a mapping population and their progeny), mapping of the sequencing reads to the reference sequence, SNP calling and filtering, SNP genotyping and imputation, followed by haplotype identification and downstream analysis. GBS has a range of applications from general marker discovery, haplotype identification, and recombination characterization to quantitative trait locus (QTL) analysis, genome-wide association studies (GWAS), and genomic selection (GS). It has already been applied to a range of plant species including: rice, maize, artichoke, and Arabidopsis thaliana. It is a promising approach which is likely to provide new and important insights into plant biology.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Jarne P, Lagoda PJL (1996) Microsatellites, from molecules to populations and back. Trends Ecol Evol 11:424–429
Arif IA, Bakir MA, Khan HA, Al Farhan AH, Al Homaidan AA, Bahkali AH, Sadoon MA, Shobrak M (2010) A brief review of molecular techniques to assess plant diversity. Int J Mol Sci 11:2079–2096
Edwards D, Forster JW, Chagné D, Batley J (2007) What are SNPs? In: Oraguzie NC, Rikkerink EHA, Gardiner SE, De Silva HN (eds) Association mapping in plants. Springer, New York, pp 41–52
Edwards D, Forster JW, Cogan NOI, Batley J, Chagné D (2007) Single nucleotide polymorphism discovery. In: Oraguzie N, Rikkerink E, Gardiner S, De Silva H (eds) Association mapping in plants. Springer, New York, pp 53–76
Chagné D, Batley J, Edwards D, Forster JW (2007) Single nucleotide polymorphism genotyping in plants. In: Oraguzie N, Rikkerink E, Gardiner S, De Silva H (eds) Association mapping in plants. Springer, New York, pp 77–94
Duran C, Edwards D, Batley J (2009) Molecular marker discovery and genetic map visualisation. In: Edwards D, Hanson D, Stajich J (eds) Applied bioinformatics. Springer, New York, pp 165–189
Hayward A, Dalton-Morgan J, Mason A, Zander M, Edwards D, Batley J (2012) SNP discovery and applications in Brassica napus. J Plant Biotechnol 39:1–12
Batley J, Edwards D (2009) Mining for single nucleotide polymorphism (SNP) and simple sequence repeat (SSR) molecular genetic markers. In: Posada D (ed) Bioinformatics for DNA sequence analysis. Humana, New York, pp 303–322
Appleby N, Edwards D, Batley J (2009) New technologies for ultra-high throughput genotyping in plants. In: Somers D, Langridge P, Gustafson J (eds) Plant genomics. Humana, New York, pp 19–40
Edwards D, Batley J, Snowdon R (2013) Accessing complex crop genomes with next-generation sequencing. Theor Appl Genet 126:1–11
Edwards D, Wang X (2012) Genome sequencing initiatives. In: Edwards D, Parkin IAP, Batley J (eds) Genetics, genomics and breeding of oilseed Brassicas. Science Publishers Inc., New Hampshire, pp 152–157
Imelfort M, Batley J, Grimmond S, Edwards D (2009) Genome sequencing approaches and successes. In: Somers D, Langridge P, Gustafson J (eds) Plant genomics. Humana, New York, pp 345–358
Edwards D, Batley J (2010) Plant genome sequencing: applications for crop improvement. Plant Biotechnol J 7:1–8
Berkman PJ, Skarshewski A, Lorenc MT, Lai K, Duran C, Ling EYS, Stiller J, Smits L, Imelfort M, Manoli S, McKenzie M, Kubalakova M, Simkova H, Batley J, Fleury D, Dolezel J, Edwards D (2011) Sequencing and assembly of low copy and genic regions of isolated Triticum aestivum chromosome arm 7DS. Plant Biotechnol J 9:768–775
Berkman PJ, Skarshewski A, Manoli S, Lorenc MT, Stiller J, Smits L, Lai K, Campbell E, Kubalakova M, Simkova H, Batley J, Dolezel J, Hernandez P, Edwards D (2012) Sequencing wheat chromosome arm 7BS delimits the 7BS/4AL translocation and reveals homoeologous gene conservation. Theor Appl Genet 124:423–432
Berkman PJ, Visendi P, Lee HC, Stiller J, Manoli S, Lorenc MT, Lai K, Batley J, Fleury D, Šimková H, Kubaláková M, Weining S, Doležel J, Edwards D (2013) Dispersion and domestication shaped the genome of bread wheat. Plant Biotechnol J 11:564–571
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J-H, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park B-S, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C, Peng C, Geng C, Koh C, Lin C, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King GJ, Bonnema G, Tang H, Wang H, Belcram H, Zhou H, Hirakawa H, Abe H, Guo H, Wang H, Jin H, Parkin IAP, Batley J, Kim J-S, Just J, Li J, Xu J, Deng J, Kim JA, Li J, Yu J, Meng J, Wang J, Min J, Poulain J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao M, Jin M, Ramchiary N, Drou N, Berkman PJ, Cai Q, Huang Q, Li R, Tabata S, Cheng S, Zhang S, Zhang S, Huang S, Sato S, Sun S, Kwon S-J, Choi S-R, Lee T-H, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li Y, Du Y, Liao Y, Lim Y, Narusaka Y, Wang Y, Wang Z, Li Z, Wang Z, Xiong Z, Zhang Z. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1040
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon SB, Baek J, Tar'an B, Millan T, Zhang X, Rosen B, Ramsay LD, Iwata A, Wang Y, Nelson W, Farmer AD, Gaur PM, Soderlund C, Penmetsa RV, Xu C, Bharti AK, He W, Winter P, Zhao S, Hane JK, Carrasquilla-Garcia N, Condie JA, Upadhyaya HD, Luo M, Singh NP, Lichtenzveig J, Gali KK, Rubio J, Nadarajan N, Thudi M, Dolezel J, Bansal KC, Xu X, Edwards D, Zhang G, Kahl G, Gil J, Singh KB, Datta SK, Jackson SA, Wang J, Cook D (2013) Draft genome sequence of kabuli chickpea (Cicer arietinum): genetic structure and breeding constraints for crop improvement. Nat Biotechnol 31:240–246
Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O'Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
IT (2013) Ion Torrent. http://www.iontorrent.com/
Imelfort M, Duran C, Batley J, Edwards D (2009) Discovering genetic polymorphisms in next-generation sequencing data. Plant Biotechnol J 7:312–317
Lorenc MT, Boskovic Z, Stiller J, Duran C, Edwards D (2012) Role of bioinformatics as a tool for oilseed Brassica species. In: Edwards D, Parkin IAP, Batley J (eds) Genetics, genomics and breeding of oilseed Brassicas. Science Publishers Inc, New Hampshire, pp 194–205
Duran C, Boskovic Z, Batley J, Edwards D (2011) Role of bioinformatics as a tool for vegetable Brassica species. In: Stiller J (ed) Vegetable Brassicas. Science Publishers, Inc., New Hampshire, pp 406–418
Edwards D (2011) Wheat bioinformatics. In: Bonjean A, Angus W, Van Ginkel M (eds) The world wheat book. Lavoisier, Paris, pp 851–875
Lee H, Lai K, Lorenc MT, Imelfort M, Duran C, Edwards D (2012) Bioinformatics tools and databases for analysis of next generation sequence data. Brief Funct Genomics 2:12–24
Berkman PJ, Lai K, Lorenc MT, Edwards D (2012) Next generation sequencing applications for wheat crop improvement. Am J Bot 99:365–371
Batley J, Edwards D (2009) Genome sequence data: management, storage, and visualization. Biotechniques 46:333–336
Duran C, Eales D, Marshall D, Imelfort M, Stiller J, Berkman PJ, Clark T, McKenzie M, Appleby N, Batley J, Basford K, Edwards D (2010) Future tools for association mapping in crop plants. Genome 53:1017–1023
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229
Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T, Dong G, Sang T, Han B (2009) High-throughput genotyping by whole-genome resequencing. Genome Res 19:1068–1076
Yang S, Yuan Y, Wang L, Li J, Wang W, Liu H, Chen J-Q, Hurst LD, Tian D (2012) Great majority of recombination events in Arabidopsis are gene conversion events. Proc Natl Acad Sci %R 101073/pnas1211827110
Gore MA, Chia J-M, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, Ware DH, Buckler ES (2009) A first-generation haplotype map of maize. Science 326:1115–1117
Scaglione D, Acquadro A, Portis E, Tirone M, Knapp SJ, Lanteri S (2012) RAD tag sequencing as a source of SNP markers in Cynara cardunculus L. BMC Genomics 13:3
Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA (2007) Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res 17:240–248
Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461:272–276
Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3:e3376
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12:499–510
Davey JW, Cezard T, Fuentes-Utrilla P, Eland C, Gharbi K, Blaxter ML (2012) Special features of RAD Sequencing data: implications for genotyping. Mol Ecol 22:3151–3164
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng Q, Liu K, Huang T, Zhou T, Jing Y, Li W, Lin Z, Buckler ES, Qian Q, Zhang Q-F, Li J, Han B (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967
Wilkening S, Tekkedil MM, Lin G, Fritsch ES, Wei W, Gagneur J, Lazinski DW, Camilli A, Steinmetz LM (2013) Genotyping 1000 yeast strains by next-generation sequencing. BMC Genomics 14:90
Sampson J, Jacobs K, Yeager M, Chanock S, Chatterjee N (2011) Efficient study design for next generation sequencing. Genet Epidemiol 35:269–277
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Lorenc MT, Hayashi S, Stiller J, Lee H, Manoli S, Ruperao P, Visendi P, Berkman PJ, Lai K, Batley J, Edwards D (2012) Discovery of single nucleotide polymorphisms in complex genomes using SGSautoSNP. Biology 1:370–382
Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18:2024–2033
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Halperin E, Stephan DA (2009) SNP imputation in association studies. Nat Biotechnol 27:349–351
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media New York
About this protocol
Cite this protocol
Golicz, A.A., Bayer, P.E., Edwards, D. (2015). Skim-Based Genotyping by Sequencing. In: Batley, J. (eds) Plant Genotyping. Methods in Molecular Biology, vol 1245. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-1966-6_19
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1966-6_19
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-1965-9
Online ISBN: 978-1-4939-1966-6
eBook Packages: Springer Protocols