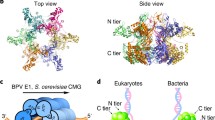
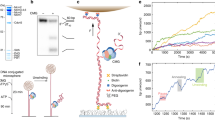
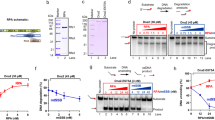
Abstract
Ring-shaped hexameric helicases play an essential role of double-stranded DNA unwinding during genome replication. The NTPase-powered unwinding activity of the hexameric helicases is required both for replication initiation and fork progression. We describe ensemble biophysical methods to measure the unwinding activity of ring-shaped helicases during fork progression using the T7 bacteriophage replicative helicase gp4A′ as a model enzyme. These assays provide insights into the stepping mechanism of translocation, active or passive mechanism of unwinding, and regulation by associated proteins such as single strand DNA binding protein, DNA polymerase, and primase enzymes.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Patel SS, Donmez I (2006) Mechanisms of helicases. J Biol Chem 281:18265–18268
Singleton MR, Dillingham MS, Wigley DB (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76:23–50
Lohman TM, Tomko EJ, Wu CG (2008) Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol 9:391–401
Egelman EH (1998) Bacterial helicases. J Struct Biol 124:123–128
Yodh JG, Schlierf M, Ha T (2010) Insight into helicase mechanism and function revealed through single-molecule approaches. Q Rev Biophys 43:185–217
Fischer CJ, Tomko EJ, Wu CG, Lohman TM (2012) Fluorescence methods to study DNA translocation and unwinding kinetics by nucleic acid motors. Methods Mol Biol 875:85–104
Costa A, Onesti S (2009) Structural biology of MCM helicases. Crit Rev Biochem Mol Biol 44:326–342
Patel SS, Picha KM (2000) Structure and function of hexameric helicases. Annu Rev Biochem 69:651–697
Rasnik I, Jeong YJ, McKinney SA, Rajagopal V, Patel SS, Ha T (2008) Branch migration enzyme as a Brownian ratchet. EMBO J 27:1727–1735
Kaplan DL, Davey MJ, O’Donnell M (2003) Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J Biol Chem 278:49171–49182
Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T (1999) Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell 99:167–177
Singleton MR, Sawaya MR, Ellenberger T, Wigley DB (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101:589–600
Thomsen ND, Berger JM (2009) Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell 139:523–534
Enemark EJ, Joshua-Tor L (2006) Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442:270–275
Liao JC, Jeong YJ, Kim DE, Patel SS, Oster G (2005) Mechanochemistry of t7 DNA helicase. J Mol Biol 350:452–475
Crampton DJ, Mukherjee S, Richardson CC (2006) DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol Cell 21:165–174
Hacker KJ, Johnson KA (1997) A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry 36:14080–14087
Ahnert P, Patel SS (1997) Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J Biol Chem 272:32267–32273
Jezewska MJ, Rajendran S, Bujalowska D, Bujalowski W (1998) Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB helicase? Fluorescence energy transfer studies. J Biol Chem 273:10515–10529
Kaplan DL (2000) The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J Mol Biol 301:285–299
Galletto R, Maillard R, Jezewska MJ, Bujalowski W (2004) Global conformation of the Escherichia coli replication factor DnaC protein in absence and presence of nucleotide cofactors. Biochemistry 43:10988–11001
Rothenberg E, Trakselis MA, Bell SD, Ha T (2007) MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J Biol Chem 282:34229–34234
Betterton MD, Julicher F (2005) Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys Rev E Stat Nonlin Soft Matter Phys 71:011904
Benkovic SJ, Valentine AM, Salinas F (2001) Replisome-mediated DNA replication. Annu Rev Biochem 70:181–208
Hamdan SM, Richardson CC (2009) Motors, switches, and contacts in the replisome. Annu Rev Biochem 78:205–243
Langston LD, Indiani C, O’Donnell M (2009) Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle 8:2686–2691
Eggleston AK, Rahim NA, Kowalczykowski SC (1996) A helicase assay based on the displacement of fluorescent, nucleic acid-binding ligands. Nucleic Acids Res 24:1179–1186
Cheng W, Hsieh J, Brendza KM, Lohman TM (2001) E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J Mol Biol 310:327–350
Picha KM, Patel SS (1998) Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+. The presence of dTTP is sufficient for hexamer formation and DNA binding. J Biol Chem 273:27315–27319
Sen D, Nandakumar D, Tang GQ, Patel SS (2012) The human mitochondrial DNA helicase TWINKLE is both an unwinding and an annealing helicase. J Biol Chem 287(18):14545–14556
Torimura M, Kurata S, Yamada K, Yokomaku T, Kamagata Y, Kanagawa T, Kurane R (2001) Fluorescence-quenching phenomenon by photoinduced electron transfer between a fluorescent dye and a nucleotide base. Anal Sci 17:155–160
Noble JE, Wang L, Cole KD, Gaigalas AK (2005) The effect of overhanging nucleotides on fluorescence properties of hybridising oligonucleotides labelled with Alexa-488 and FAM fluorophores. Biophys Chem 113:255–263
Toseland CP, Webb MR (2010) Fluorescence tools to measure helicase activity in real time. Methods 51:259–268
Xi XG, Deprez E (2010) Monitoring helicase-catalyzed DNA unwinding by fluorescence anisotropy and fluorescence cross-correlation spectroscopy. Methods 51:289–294
Dou SX, Xi XG (2010) Fluorometric assays for characterizing DNA helicases. Methods 51:295–302
Donmez I, Patel SS (2008) Coupling of DNA unwinding to nucleotide hydrolysis in a ring-shaped helicase. EMBO J 27:1718–1726
Sun B, Johnson DS, Patel G, Smith BY, Pandey M, Patel SS, Wang MD (2011) ATP-induced helicase slippage reveals highly coordinated subunits. Nature 478:132–135
Manosas M, Spiering MM, Zhuang Z, Benkovic SJ, Croquette V (2009) Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome. Nat Chem Biol 5:904–912
Eoff RL, Raney KD (2006) Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nat Struct Mol Biol 13:242–249
Lucius AL, Maluf NK, Fischer CJ, Lohman TM (2003) General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys J 85:2224–2239
Levin MK, Hingorani MM, Holmes RM, Patel SS, Carson JH (2009) Model-based global analysis of heterogeneous experimental data using gfit. Methods Mol Biol 500:335–359
Jeong YJ, Levin MK, Patel SS (2004) The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc Natl Acad Sci U S A 101:7264–7269
Breslauer KJ, Frank R, Blocker H, Marky LA (1986) Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A 83:3746–3750
Johnson DS, Bai L, Smith BY, Patel SS, Wang MD (2007) Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell 129:1299–1309
Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V (2007) Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc Natl Acad Sci U S A 104:19790–19795
Ribeck N, Kaplan DL, Bruck I, Saleh OA (2010) DnaB helicase activity is modulated by DNA geometry and force. Biophys J 99:2170–2179
Rajagopal V, Patel SS (2008) Single strand binding proteins increase the processivity of DNA unwinding by the hepatitis C virus helicase. J Mol Biol 376:69–79
Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol 43:289–318
Frick DN, Richardson CC (2001) DNA primases. Annu Rev Biochem 70:39–80
Kusakabe T, Baradaran K, Lee J, Richardson CC (1998) Roles of the helicase and primase domain of the gene 4 protein of bacteriophage T7 in accessing the primase recognition site. EMBO J 17:1542–1552
Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS (2009) Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature 462:940–943
Yuzhakov A, Kelman Z, O’Donnell M (1999) Trading places on DNA—a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell 96:153–163
Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM (2006) DNA primase acts as a molecular brake in DNA replication. Nature 439:621–624
Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS (2005) DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature 435:370–373
Manosas M, Spiering MM, Ding F, Croquette V, Benkovic SJ (2012) Collaborative coupling between polymerase and helicase for leading-strand synthesis. Nucleic Acids Res 40(13):6187–6198
Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I Jr, Pyle AM, Bustamante C (2006) RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439:105–108
Graham BW, Schauer GD, Leuba SH, Trakselis MA (2011) Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res 39:6585–6595
Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, Lilley DM, Ha T (2007) Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the holliday junction. Science 318:279–283
Zhou R, Kozlov AG, Roy R, Zhang J, Korolev S, Lohman TM, Ha T (2011) SSB functions as a sliding platform that migrates on DNA via reptation. Cell 146:222–232
Acknowledgements
We thank former and current Patel Lab members for developing and testing the methods and models described in this review. This work was supported by National Institute of Health (NIH) grant GM55310.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media New York
About this chapter
Cite this chapter
Nandakumar, D., Patel, S.S. (2013). Helicase Unwinding at the Replication Fork. In: Allewell, N., Narhi, L., Rayment, I. (eds) Molecular Biophysics for the Life Sciences. Biophysics for the Life Sciences, vol 6. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-8548-3_9
Download citation
DOI: https://doi.org/10.1007/978-1-4614-8548-3_9
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-8547-6
Online ISBN: 978-1-4614-8548-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)