Abstract
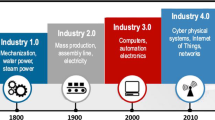
The bio-manufacturing industry, along with other process industries, now has the opportunity to be engaged in the latest industrial revolution, also known as Industry 4.0. To successfully accomplish this, a physical-to-digital-to-physical information loop should be carefully developed. One way to achieve this is, for example, through the implementation of digital twins (DTs), which are virtual copies of the processes. Therefore, in this paper, the focus is on understanding the needs and challenges faced by the bio-manufacturing industry when dealing with this digitalized paradigm. To do so, two major building blocks of a DT, data and models, are highlighted and discussed. Hence, firstly, data and their characteristics and collection strategies are examined as well as new methods and tools for data processing. Secondly, modelling approaches and their potential of being used in DTs are reviewed. Finally, we share our vision with regard to the use of DTs in the bio-manufacturing industry aiming at bringing the DT a step closer to its full potential and realization.
Graphical Abstract

Carina L. Gargalo and Simoneta Caþo de las Heras are both first authors.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- AI:
-
Artificial intelligence
- AIC:
-
Akaike’s information criterion
- ANN:
-
Artificial neural networks
- BCO:
-
BioCompute Object
- BIC:
-
Bayesian information criterion
- BSM:
-
Benchmark simulation models
- CAPE-OPEN:
-
Next-generation computer-aided process engineering open simulation environment
- CFD:
-
Computational fluid dynamics
- CM:
-
Compartmental models
- CQA:
-
Critical quality attributes
- DCS:
-
Distributed control systems
- DEXPI:
-
Data exchange in the process industry
- DT:
-
Digital twins
- FPM:
-
First-principles models
- GC/MS:
-
Gas chromatography/mass spectrometry
- GLP:
-
Good laboratory practice
- GMP:
-
Good modelling practices
- HPC:
-
High-performance computing
- IoT:
-
Internet of Things
- ISO:
-
International Organization for Standardization
- LCA:
-
Multilinear regression
- MIR:
-
Mid infrared
- OPC:
-
Open platform communications
- ML:
-
Machine learning
- MPC:
-
Model predictive control
- MVA:
-
Multivariate data analysis
- PAT:
-
Process analytical technology
- PCA:
-
Principal component analysis
- PID:
-
Proportional-integral-derivative
- PLC:
-
Programmable logic controllers
- PLS:
-
Partial least squares regression
- QbD:
-
Quality by design
- RTU:
-
Remote terminal units
- SAX:
-
Symbolic aggregate proximity
- SCADA:
-
Supervisory control and data acquisition
- SSE:
-
Sum of squared errors
- SVM:
-
Support vector machines
- UV:
-
Ultraviolet
- WWTP:
-
Wastewater treatment plants
- XML:
-
Extensible markup language
References
Sniderman B, Mahto M, Cotteleer MJ (2016) Industry 4.0 and manufacturing ecosystems: exploring the world of connected enterprises. Deloitte Consulting
Kagermann H, Wahlster W, Helbig J (2013) Securing the future of german manufacturing industry: recommendations for implementing the strategic initiative industrie 4.0. Technical report 0
Oliveira AL (2019) Biotechnology, big data and artificial intelligence. Biotechnol J 14(8):1800613
Deloitte. Digital Thread for Additive Manufacturing (DTAM)
Fraunhofer Austria Research GmbH. Innovative Solutions for the Today of Tomorrow
Zhang M, Nee Fei Tao AYC (2019) Background and concept of digital twin. In: Digital twin driven smart manufacturing. Academic Press, pp 3–28
Garetti M, Rosa P, Terzi S (2012) Life cycle simulation for the design of product-service systems. Comput Indust 63(4):361–369
Kritzinger W, Karner M, Traar G, Henjes J, Sihn W (2018) Digital twin in manufacturing: a categorical literature review and classification. IFAC-PapersOnLine 51(11):1016–1022
Negri E, Fumagalli L, Macchi M (2017) A review of the roles of digital twin in CPS-based production systems. Proc Manuf 11:939–948
Hermann M, Pentek T, Otto B (2016) Design principles for industrie 4.0 scenarios. In: 2016 49th Hawaii international conference on system sciences (HICSS). IEEE, pp 3928–3937
Tao F, Qi Q, Wang L, Nee AYC (2019) Digital twins and cyber–physical systems toward smart manufacturing and industry 4.0: correlation and comparison. Engineering 5(4):653–661
O’Donovan P, Leahy K, Bruton K, O’Sullivan DTJ (2015) An industrial big data pipeline for data-driven analytics maintenance applications in large-scale smart manufacturing facilities. J Big Data 2(1):25
Tao F, Qi Q, Liu A, Kusiak A (2018) Data-driven smart manufacturing. J Manuf Syst 48:157–169
Rosen R, VON Wichert G, Lo G, Bettenhausen KD (2015) About the importance of autonomy and digital twins for the future of manufacturing. IFAC-PapersOnLine 48(3):567–572
Wright L, Davidson S (2020) How to tell the difference between a model and a digital twin. Adv Model Simulat Eng Sci 7(1):13
Baur D, Angelo J, Chollangi Ss, Müller-Späth T, Xu X, Ghose S, Li ZJ, Morbidelli M (2019) Model-assisted process characterization and validation for a continuous two-column protein A capture process. Biotechnol Bioeng 116(1):87–98
Möller J, Kuchemüller KB, Steinmetz T, Koopmann KS, Pörtner R (2019) Model-assisted design of experiments as a concept for knowledge-based bioprocess development. Bioprocess Biosyst Eng 42(5):867–882
Warshaw L, Cotteleer M (2017) Industry 4.0 and the digital twin. Deloite University Press
Holdowsky J, Mahto M, Raynor ME, Cotteleer M (2015) Inside the internet of things (iot). Retrieved 5 Apr 2016
Madni AM, Madni CC, Lucero SD (2019) Leveraging digital twin technology in model-based systems engineering. Systems 7(1):7
Madni AM, Sievers M (2018) Model-based systems engineering: motivation, current status, and research opportunities. Syst Eng 21(3):172–190
Madni AM, Sievers M (2017) Model-based systems engineering: motivation, current status, and needed advances. Disciplin Converg Syst Eng Res:311–325
Lim KYH, Zheng P, Chen CH (2020) A state-of-the-art survey of digital twin: techniques, engineering product lifecycle management and business innovation perspectives. J Intell Manuf 31(6):1313–1337
Boschert S, Rosen R (2016) Digital twin-the simulation aspect. Mechatronic futures: challenges and solutions for mechatronic systems and their designers, pp 59–74
Swedberg C (2018) Digital twins bring value to big RFID and IoT data. https://www.rfidjournal.com/digital-twins-bring-value-to-big-rfid-and-iot-data-2
Menard S (2017) 3 ways digital twins are going to help improve oil and gas maintenance and operations. https://www.linkedin.com/pulse/3-ways-digitaltwins-going-help-improve-oil-gas-sophie-menard
Science Service Dr (2017) Hempel Digital Health Network. Healthcare solution testing for future|Digital Twins in healthcare. https://www.dr-hempel-network.com/digital-health-technolgy/digital-twins-in-healthcare/
DNV.GL. WINDGEMINI DIGITAL TWIN: Data driven insights to reduce costs, extend life and maximise production. https://www.dnvgl.com/power-renewables/services/data-analytics/windgemini/?utm_campaign=wind&utm_source=google&utm_medium=cpc&utm_content=250560941230&utm_term=wind%20turbine%20digital%20twin&gclid=Cj0KCQjw3ZX4BRDmARIsAFYh7ZKDzHi57l1WoqfTZDz6VL6yfICGSef_mvHLkZOl90uzgcSgPvSxVMYaAoT1EALw_wcB
Dassault Systèmes (2018) Meet virtual Singapore, the city’s 3D digital twin. https://govinsider.asia/digitalgov/meet-virtual-singapore-citys-3d-digital-twin/
Narayanan H, Luna MF, von Stosch M, Cruz Bournazou MN, Polotti G, Morbidelli M, Butté A, Sokolov M (2019) Bioprocessing in the digital age: the role of process models. Biotechnol J:1900172
Endo I, Nagamune T (1987) A database system for fermentation processes. Bioprocess Eng 2(3):111–114
FDA. FDA’s regulation of plant and animal biotechnology products
Markarian J (2018) Modernizing pharma manufacturing. Pharm Technol 42(4):20–25
Mercier SM, Diepenbroek B, Wijffels RH, Streefland M (2014) Multivariate PAT solutions for biopharmaceutical cultivation: current progress and limitations. Trends Biotechnol 32(6):329–336
Simon LL, Pataki H, Marosi G, Meemken F, Hungerbühler K, Baiker A, Tummala S, Glennon B, Kuentz M, Steele G et al (2015) Assessment of recent process analytical technology (pat) trends: a multiauthor review. Org Process Res Dev 19(1):3–62
Teixeira AP, Oliveira R, Alves PM, Carrondo MJT (2009) Advances in on-line monitoring and control of mammalian cell cultures: supporting the PAT initiative. Biotechnol Adv 27(6):726–732
Kroll P, Hofer A, Ulonska S, Kager J, Herwig C (2017) Model-based methods in the biopharmaceutical process lifecycle. Pharm Res 34(12):2596–2613
Venkatasubramanian V (2019) The promise of artificial intelligence in chemical engineering: is it here, finally. AICHE J 65(2):466–478
Glassey J, Von Stosch M (2018) Hybrid modeling in process industries. CRC Press
von Stosch M, Davy S, Francois K, Galvanauskas V, Hamelink J, Luebbert A, Mayer M, Oliveira R, O’Kennedy R, Rice P, Glassey JA (2014) Hybrid modeling for quality by design and PAT – benefits and challenges of applications in biopharmaceutical industry. Biotechnol J 9:719–726
Reis MS, Gins G (2017) Industrial process monitoring in the big data/industry 4.0 era: from detection, to diagnosis, to prognosis. PRO 5(3):35
Steinwandter V, Borchert D, Herwig C (2019) Data science tools and applications on the way to Pharma 4.0. Drug Discov Today 24(9):1795–1805
Freesense. https://www.freesense.dk
Particletech solutions. https://particletech.dk/particletechsolution/
Lawton JR, Martinez FA, Burks C (1989) Overview of the limb database. Nucleic Acids Res 17(15):5885–5889
Biotrack product database. https://biotrackproductdatabase.oecd.org
Biechele P, Busse C, Solle D, Scheper T, Reardon K (2015) Sensor systems for bioprocess monitoring. Eng Life Sci 15(5):469–488
Zimmermann R, Fiabane L, Gasteuil Y, Volk R, Pinton J (2013) Measuring lagrangian accelerations using an instrumented particle. Phys Scr 2013(T155):014063
Landgrebe D, Haake C, Höpfner T, Beutel S, Hitzmann B, Scheper T, Rhiel M, Reardon KF (2010) On-line infrared spectroscopy for bioprocess monitoring. Appl Microbiol Biotechnol 88(1):11–22
Kadlec P, Gabrys B, Strandt S (2009) Data-driven soft sensors in the process industry. Comput Chem Eng 33(4):795–814
Pohlscheidt M, Charaniya S, Bork C, Jenzsch M, Noetzel TL, Luebbert A (2009) Bioprocess and fermentation monitoring. Encycl Indust Biotechnol Bioprocess Biosep Cell Technol:1469–1491
Gopakumar V, Tiwari S, Rahman I (2018) A deep learning based data driven soft sensor for bioprocesses. Biochem Eng J 136:28–39
Spann R, Roca C, Kold D, Lantz AE, Gernaey KV, Sin G (2018) A probabilistic model-based soft sensor to monitor lactic acid bacteria fermentations. Biochem Eng J 135:49–60
Thürlimann CM, Dürrenmatt DJ, Villez K (2018) Soft-sensing with qualitative trend analysis for wastewater treatment plant control. Control Eng Pract 70:121–133
Qi Q, Tao F (2018) Digital twin and big data towards smart manufacturing and industry 4.0: 360 degree comparison. IEEE Access 6:3585–3593
Chen M, Mao S, Liu Y (2014) Big data: a survey. Mob Networks Appl 19(2):171–209
Boiarkina I, Depree N, Prince-Pike A, Yu W, Wilson DI, Young BR (2018) Using big data in industrial milk powder process systems. In: Computer aided chemical engineering, vol 44. Elsevier, pp 2293–2298
Data warehouse, data lake and database definition. https://blogs.oracle.com/bigdata/data-lake-database-data-warehouse-difference
Charaniya S, Hu W, Karypis G (2008) Mining bioprocess data: opportunities and challenges. Trends Biotechnol 26(12):690–699
Mercier SM, Diepenbroek B, Dalm M, Wijffels RH, Streefland M (2013) Multivariate data analysis as a pat tool for early bioprocess development data. J Biotechnol 167(3):262–270
Al-Maskari S, Li X, Liu Q (2014) An effective approach to handling noise and drift in electronic noses. In: Australasian database conference. Springer, Berlin, pp 223–230
Goodner KL, Dreher JG, Rouseff RL (2001) The dangers of creating false classifications due to noise in electronic nose and similar multivariate analyses. Sensors Actuators B Chem 80(3):261–266
Xie W, Li C, Wu Y, Zhang P (2019) A bayesian nonparametric framework for uncertainty quantification in simulation. arXiv preprint arXiv:1910.03766
Gupta SK (2012) Use of Bayesian statistics in drug development: advantages and challenges. Int J Appl Basic Med Res 2(1):3–6
Tabora JE, Gonzalez FL, Tom JW (2019) Bayesian probabilistic modeling in pharmaceutical process development. AICHE J 65(11):e16744
García-Muñoz S, Luciani CV, Vaidyaraman S, Seibert KD (2015) Definition of design spaces using mechanistic models and geometric projections of probability maps. Org Process Res Dev 19(8):1012–1023
Richard X, Laird C, Vaidyaraman S, García-Muñoz S (2017) An optimization-based framework to define the probabilistic design space of pharmaceutical processes with model uncertainty. In: Computing and systems technology division 2017 – Core programming area at the 2017 aiche annual meeting, vol 2017. AIChE, pp 610–622
Albrecht J (2013) Estimating reaction model parameter uncertainty with Markov chain Monte Carlo. Comput Chem Eng 48:14–28
Rathore AS, Bhushan A, Hadpe S (2011) Chemometrics applications in biotech processes: a review. Biotechnol Prog 27(2):307–315
Turitsyn SK, Prilepsky JE, Le ST, Wahls S, Frumin LL, Kamalian M, Derevyanko SA (2017) Nonlinear fourier transform for optical data processing and transmission: advances and perspectives. Optica 4(3):307–322
Notaristefano A, Chicco G, Piglione F (2013) Data size reduction with symbolic aggregate approximation for electrical load pattern grouping. IET Gen Trans Distrib 7(2):108–117
Keogh E, Chakrabarti K, Pazzani M, Mehrotra S (2001) Locally adaptive dimensionality reduction for indexing large time series databases. In: Proceedings of the 2001 ACM SIGMOD international conference on Management of data, pp 151–162
Cordella CB (2012) Pca: the basic building block of chemometrics. Anal Chem 154
Roggo Y, Chalus P, Maurer L, Lema-Martinez C, Edmond A, Jent N (2007) A review of near infrared spectroscopy and chemometrics in pharmaceutical technologies. J Pharm Biomed Anal 44(3):683–700
Desai K, Badhe Y, Tambe SS, Kulkarni BD (2006) Soft-sensor development for fed-batch bioreactors using support vector regression. Biochem Eng J 27(3):225–239
Dieterle F, Busche S, Gauglitz G (2004) Different approaches to multivariate calibration of nonlinear sensor data. Anal Bioanal Chem 380(3):383–396
Taib MN, Andres R, Narayanaswamy R (1996) Extending the response range of an optical fibre ph sensor using an artificial neural network. Anal Chim Acta 330(1):31–40
Corominas L, Garrido-Baserba M, Villez K, Olsson G, Cortés U, Poch M (2018) Transforming data into knowledge for improved wastewater treatment operation: a critical review of techniques. Environ Model Softw 106:89–103
Lopez PC, Udugama IA, Thomsen ST, Roslander C, Junicke H, Mauricio-Iglesias M, Gernaey KV (2020) Towards a digital twin: a hybrid data-driven and mechanistic digital shadow to forecast the evolution of lignocellulosic fermentation. Biofuels Bioprod Biorefin
Elsevier’s scopus, the largest abstract and citation database of peer-reviewed literature. https://www.scopus.com/
Kell DB, Sonnleitner B (1995) Gmp – good modelling practice: an essential component of good manufacturing practice. Trends Biotechnol 13(11):481–492
Mears L, Stocks SM, Albaek MO, Sin G, Gernaey KV (2017) Mechanistic fermentation models for process design, monitoring, and control. Trends Biotechnol 35(10):914–924
Kroll P, Hofer A, Stelzer IV, Herwig C (2017) Workflow to set up substantial target-oriented mechanistic process models in bioprocess engineering. Process Biochem 62:24–36
Gernaey KV, Lantz AE, Tufvesson P, Woodley JM, Sin G (2010) Application of mechanistic models to fermentation and biocatalysis for next-generation processes. Trends Biotechnol 28(7):346–354
Sonnleitner B, Käppeli O (1986) Growth of saccharomyces cerevisiae is controlled by its limited respiratory capacity: formulation and verification of a hypothesis. Biotechnol Bioeng 28(6):927–937
Song H, Jang SH, Park JM, Lee SY (2008) Modeling of batch fermentation kinetics for succinic acid production by mannheimia succiniciproducens. Biochem Eng J 40(1):107–115
Sin G, Ödman P, Petersen N, Lantz AE, Gernaey KV (2008) Matrix notation for efficient development of first-principles models within pat applications: integrated modeling of antibiotic production with streptomyces coelicolor. Biotechnol Bioeng 101(1):153–171
Abt V, Barz T, Cruz-Bournazou MN, Herwig C, Kroll P, Möller J, Pörtner R, Schenkendorf R (2018) Model-based tools for optimal experiments in bioprocess engineering. Curr Opin Chem Eng 22:244–252
Lübbert A, Simutis R (1994) Using measurement data in bioprocess modelling and control. Trends Biotechnol 12(8):304–311
Boyd S, Vandenberghe L (2018) Introduction to applied linear algebra: vectors, matrices, and least squares. Cambridge University Press
Burnham KP, Anderson DR (2002) A practical information-theoretic approach. Model selection and multimodel inference, 2nd edn. Springer, New York
Cozad A, Sahinidis NV, Miller DC (2014) Learning surrogate models for simulation-based optimization. AICHE J 60(6):2211–2227
Ghojogh B, Crowley M (2019) The theory behind overfitting, cross validation, regularization, bagging, and boosting: Tutorial. p 23
Williams BA, Cremaschi S (2019) Surrogate model selection for design space approximation and surrogatebased optimization. In: Proceedings of the 9th international conference on foundations of computer-aided process design, vol 47 of Computer Aided Chemical Engineering, pp 353–358
Bishop CM (2006) Pattern recognition and machine learning (information science and statistics). Springer, Berlin
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19(6):716–723
Mallows CL (1973) Some comments on cp. Technometrics 15(4):661–675
Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6(2):461–464
Hannan EJ, Quinn BG (1979) The determination of the order of an autoregression. J R Stat Soc Ser B (Methodol) 41(2):190–195
Jones M, Forero-Hernandez H, Zubov A, Sarup B, Sin G (2018) Superstructure optimization of oleochemical processes with surrogate models. In Proceedings of the 13th international symposium on process systems engineering – PSE 2018, volume 44 of Computer Aided Chemical Engineering, pp 277–282
Al R, Behera CR, Zubov A, Gernaey KV, Sin G (2019) Meta-modeling based efficient global sensitivity analysis for wastewater treatment plants – an application to the bsm2 model. Comput Chem Eng 127:233–246
Davis SE, Cremaschi S, Eden MR (2018) Efficient surrogate model development: impact of sample size and underlying model dimensions, vol 44, pp 979–984
Garud SS, Karimi IA, Kraft M (2017) Design of computer experiments: a review. Comput Chem Eng 106:71–95
Wilson ZT, Sahinidis NV (2017) The Alamo approach to machine learning. Comput Chem Eng 106:785–795
Alizadeh R, Jia L, Nellippallil AB, Wang G, Hao J, Allen JK, Mistree F (2019) Ensemble of surrogates and cross-validation for rapid and accurate predictions using small data sets. Artif Intell Eng Des Anal Manuf 33(4):484–501
Esche E, Weigert J, Budiarto T, Hoffmann C, Repke J-U (2019) Optimization under uncertainty based on a data-driven model for a chloralkali electrolyzer cell. In: 29th European symposium on computer aided process engineering, volume 46 of Computer Aided Chemical Engineering. Elsevier, pp 577–582
Viana FAC, Haftka RT, Steffen V (2009) Multiple surrogates: how cross-validation errors can help us to obtain the best predictor. Struct Multidiscip Optim 39(4):439–457
McBride K, Sundmacher K (2019) Overview of surrogate modeling in chemical process engineering. Chem Ingen Tech 91(3):228–239
Tajsoleiman T (2018) Automating experimentation in miniaturized reactors
Nauha EK, Kálal Z, Ali JM, Alopaeus V (2018) Compartmental modeling of large stirred tank bioreactors with high gas volume fractions. Chem Eng J 334:2319–2334
Spann R, Gernaey KV, Sin G (2019) A compartment model for risk-based monitoring of lactic acid bacteria cultivations. Biochem Eng J 151:107293
Öner M, Stocks SM, Sin G (2020) Comprehensive sensitivity analysis and process risk assessment of large scale pharmaceutical crystallization processes. Comput Chem Eng 135:106746
Noorman HJ, Heijnen JJ (2017) Biochemical engineering’s grand adventure. Chem Eng Sci 170:677–693
Nielsen RF, Kermani NA, la Cour Freiesleben L, Gernaey KV, Mansouri SS (2019) Novel strategies for predictive particle monitoring and control using advanced image analysis. In: 29th European Symposium on Computer Aided Process Engineering, 46:1435–1440
Nielsen RF, Nazemzadeh N, Sillesen LW, Andersson MP, Gernaey KV, Mansouri SS (2020) Hybrid machine learning assisted modelling framework for particle processes. Comput Chem Eng 140:106916
Eikens B, Karim MN, Simon L (1999) Neural networks and first principle models for bioprocesses. IFAC Proc 32(2):6974–6979
Gao Y, Kipling K, Glassey J, Willis M, Montague G, Zhou Y, Titchener-Hooker NJ (2010) Application of agent-based system for bioprocess description and process improvement. Biotechnol Prog 26(3):706–716
Downs JJ, Vogel EF (1993) A plant-wide industrial process control problem. Comput Chem Eng 17(3):245–255
Gernaey KV, Jeppsson U, Vanrolleghem PA, Copp JB (2014) Benchmarking of control strategies for wastewater treatment plants. IWA Publishing
Jeppsson U, Pons M-N, Nopens I, Alex J, Copp JB, Gernaey KV, Rosén C, Steyer J-P, Vanrolleghem PA (2007) Benchmark simulation model no 2: general protocol and exploratory case studies. Water Sci Technol 56(8):67–78
Nopens I, Benedetti L, Jeppsson U, Pons M-N, Alex J, Copp JB, Gernaey KV, Rosen C, Steyer J-P, Vanrolleghem PA (2010) Benchmark simulation model no 2: finalisation of plant layout and default control strategy. Water Sci Technol 62(9):1967–1974
Flores-Alsina X, Corominas L, Snip L, Vanrolleghem PA (2011) Including greenhouse gas emissions during benchmarking of wastewater treatment plant control strategies. Water Res 45(16):4700–4710
Ochoa S, Wozny G, Repke J-U (2010) Plantwide optimizing control of a continuous bioethanol production process. J Process Control 20(9):983–998
Feldman H, Flores-Alsina X, Ramin P, Kjellberg K, Jeppsson U, Batstone DJ, Gernaey KV (2017) Modelling an industrial anaerobic granular reactor using a multi-scale approach. Water Res 126:488–500
Lopez PC, Feldman H, Mauricio-Iglesias M, Junicke H, Huusom JK, Gernaey KV (2019) Benchmarking real-time monitoring strategies for ethanol production from lignocellulosic biomass. Biomass Bioenergy 127:105296. 73–86
Udugama IA, Gernaey KV, Taube MA, Bayer C (2020) A novel use for an old problem: the Tennessee Eastman challenge process as an activating teaching tool. Educ Chem Eng 30:20–31
Ricardez-Sandoval LA, Douglas PL, Budman HM (2011) A methodology for the simultaneous design and control of large-scale systems under process parameter uncertainty. Comput Chem Eng 35(2):307–318
Montes F, Gernaey KV, Sin G (2018) Dynamic plantwide modeling, uncertainty, and sensitivity analysis of a pharmaceutical upstream synthesis: ibuprofen case study. Ind Eng Chem Res 57(30):10026–10037
Ricker NL (1996) Decentralized control of the Tennessee Eastman challenge process. J Process Control 6(4):205–221
Kulkarni A, Jayaraman VK, Kulkarni BD (2005) Knowledge incorporated support vector machines to detect faults in Tennessee Eastman process. Comput Chem Eng 29(10):2128–2133
Mears L, Stocks SM, Sin G, Gernaey KV (2017) A review of control strategies for manipulating the feed rate in fed-batch fermentation processes. J Biotechnol 245:34–46
A. Udugama I, Munir MT, Kirkpatrick R, Young BR, Yu W (2018) Side draw control design for a high purity multi-component distillation column. ISA Trans 76:167–177
Bähner FD, Santacoloma PA, Huusom JK (2019) Assessment of the plantwide control structure in a pectin production plant. IFAC-PapersOnLine 52(1):251–256
Lopez PC, Udugama IA, Thomsen ST, Roslander C, Junicke H, Mauricio-Iglesias M, Gernaey KV (2020) Towards a digital twin: a hybrid data-driven and mechanistic digital shadow to forecast the evolution of lignocellulosic fermentation. In: Biofuels, bioproducts and biorefining. Wiley
Zhang H (2009) Software sensors and their applications in bioprocess. pp 25–56
Nakhaeinejad M, Bryant MD (2011) Observability analysis for model-based fault detection and sensor selection in induction motors. Meas Sci Technol 22(7):075202
Gernaey KV, Cervera-Padrell AE, Woodley JM (2012) A perspective on PSE in pharmaceutical process development and innovation. Comput Chem Eng 42:15–29
Rathore AS, Bhambure R, Ghare V (2010) Process analytical technology (PAT) for biopharmaceutical products. Anal Bioanal Chem 398(1):137–154
Lopez PC, Feldman H, Mauricio-Iglesias M, Junicke H, Huusom JK, Gernaey KV (2019) Benchmarking real-time monitoring strategies for ethanol production from lignocellulosic biomass. Biomass Bioenergy 127:73–86. 105296
Tobyn M, Ferreira AP, Morris C, Menezes JC (2018) The preeminence of multivariate data analysis as a statistical data analysis technique in Pharmaceutical R&D and manufacturing. In: Multivariate analysis in the pharmaceutical industry. Elsevier, pp 3–12
Kourti T, MacGregor JF (1995) Process analysis, monitoring and diagnosis, using multivariate projection methods. Chemom Intell Lab Syst 28(1):3–21
Udugama I, Gargalo L, Yamashita Y, Taube MA, Palazoglu A, Young BR, Gernaey KV, Kulahci M, Bayer C (2020) The role of big data in industrial (bio)chemical process operations. Ind Eng Chem Res
Morari M, H. Lee J (1999) Model predictive control: past, present and future. Comput Chem Eng 23(4–5):667–682
Liu C, Gong Z, Shen B, Feng E (2013) Modelling and optimal control for a fed-batch fermentation process. Appl Math Model 37(3):695–706
Gomes J, Chopda VR, Rathore AS (2015) Integrating systems analysis and control for implementing process analytical technology in bioprocess development. J Chem Technol Biotechnol 90(4):583–589
Sommeregger W, Sissolak B, Kandra K, von Stosch M, Mayer M, Striedner G (2017) Quality by control: towards model predictive control of mammalian cell culture bioprocesses. Biotechnol J 12(7):1600546
Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, Van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, Van Der Lei J, Van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Comment BM (2016) The fair guiding principles for scientific data management and stewardship. Sci Data 3(1):160018
Gernaey KV, Rosen C, Batstone DJ, Alex J (2006) Efficient modelling necessitates standards for model documentation and exchange. Water Sci Technol 53(1):277–285
Simonyan V, Goecks J, Mazumder R (2017) Biocompute objects – a step towards evaluation and validation of biomedical scientific computations. PDA J Pharm Sci Technol 71(2):136–146
Home page – opc foundation. https://opcfoundation.org/. Accessed 28 Apr 2020
The Cape-Open Laboratories Network. http://www.colan.org/. Accessed 28 Apr 2020
Dexpi – data exchange in the process industry. https://dexpi.org/. Accessed 28 Apr 2020
Protocol buffers. https://developers.google.com/protocol-buffers. Accessed 28 Apr 2020
gRPC – a high-performance, open source universal rpc framework. https://grpc.io/. Accessed 28 Apr 2020
Ladner RE (1975) On the structure of polynomial time reducibility. JACM 22(1):155–171
Acknowledgments
Financial support of the Novo Nordisk Foundation in the frame of the “Accelerated Innovation in Manufacturing Biologics” (AIMBio) project (grant number NNF19SA0035474) is gratefully acknowledged. In addition, the Technical University of Denmark (DTU) is acknowledged for the financial support of the PhD project of Simoneta Caño de las Heras.
Greater Copenhagen Food Innovation project (CPH-FOOD) is acknowledged for financially supporting the postdoc position of Mark Jones.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Gargalo, C.L. et al. (2020). Towards the Development of Digital Twins for the Bio-manufacturing Industry. In: Herwig, C., Pörtner, R., Möller, J. (eds) Digital Twins. Advances in Biochemical Engineering/Biotechnology, vol 176. Springer, Cham. https://doi.org/10.1007/10_2020_142
Download citation
DOI: https://doi.org/10.1007/10_2020_142
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-71659-2
Online ISBN: 978-3-030-71660-8
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)