Abstract
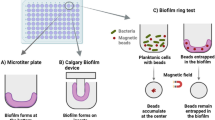
Three dynamic models for the investigation of in vitro biofilm formation are described in this chapter. In the 6-well plate assay presented here, the placing of the plate on a rotating platform provides shear, thereby making the system dynamic with respect to the static microtiter assay.
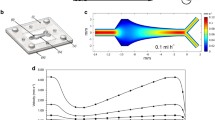
The second reported model, especially suitable for harvesting high amounts of cells for transcriptomic or proteomic investigations, is based on numerous glass beads placed in a flask incubated with shaking on a rotating platform, thus increasing the surface area for biofilm formation. Finally, the flow-cell system, that is the driving model for elucidating the biofilm-forming process in vitro as well as the biofilm tolerance towards antibiotics and host defense components, is illustrated here.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Christensen GD, Simpson WA, Younger JJ et al (1985) Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol 22:996–1006
O’Toole GA, Kolter R (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461
Ceri H, Olson ME, Stremick C et al (1999) The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776
Lee B, Haagensen JA, Ciofu O et al (2005) Heterogeneity of biofilms formed by nonmucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J Clin Microbiol 43:5247–5255
Hengzhuang W, Wu H, Ciofu O et al (2011) Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 55:4469–4474
Christensen BB, Sternberg C, Andersen JB et al (1999) Molecular tools for study of biofilm physiology. Methods Enzymol 310: 20–42
Anderl JN, Franklin MJ, Stewart PS (2000) Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother 44:1818–1824
Goeres DM, Hamilton MA, Beck NA et al (2009) A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat Protoc 4: 783–788
Zelver N, Hamilton M, Pitts B et al (1999) Measuring antimicrobial effects on biofilm bacteria: from laboratory to field. Methods Enzymol 310:608–628
Wolfaardt GM, Lawrence JR, Robarts RD et al (1994) Multicellular organization in a degradative biofilm community. Appl Environ Microbiol 60:434–446
Klausen M, Heydorn A, Ragas P et al (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48:1511–1524
Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T (2003) Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol 50:61–68
Sauer K, Camper AK, Ehrlich GD et al (2002) Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154
Stoodley P, Sauer K, Davies DG, Costerton JW (2002) Biofilms as complex differentiated communities. Annu Rev Microbiol 56: 187–209
Bjarnsholt T, Jensen PØ, Burmølle M et al (2005) Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373–383
Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T (2008) Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 68: 223–240
Alhede M, Bjarnsholt T, Jensen PØ et al (2009) Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155:3500–3508
Jensen PO, Bjarnsholt T, Phipps R et al (2007) Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153: 1329–1338
van Gennip M, Christensen LD, Alhede M et al (2009) Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS 117: 537–546
Pamp SJ, Tolker-Nielsen T (2007) Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J Bacteriol 189:2531–2539
Peters BM, Ovchinnikova ES, Krom BP et al (2012) Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology 158: 2975–2986
Bjerkan G, Witso E, Bergh K (2009) Sonication is superior to scraping for retrieval of bacteria in biofilm on titanium and steel surfaces in vitro. Acta Orthop 80:245–250
Fux CA, Wilson S, Stoodley P (2004) Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J Bacteriol 186:4486–4491
Brady RA, O’May GA, Leid JG et al (2011) Resolution of Staphylococcus aureus biofilm infection using vaccination and antibiotic treatment. Infect Immun 79:1797–1803
Abrámoff MD, Magalhâes PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Int 11:36–41
Beyenal H, Donovan C, Lewandowski Z, Harkin G (2004) Three-dimensional biofilm structure quantification. J Microbiol Methods 59:395–413
Heydorn A, Nielsen AT, Hentzer M et al (2000) Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407
Daims H, Lucker S, Wagner M (2006) Daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol 8:200–213
Southey-Pillig CJ, Davies DG, Sauer K (2005) Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol 187:8114–8126
Crusz SA, Popat R, Rybtke MT et al (2012) Bursting the bubble on bacterial biofilms: a flow cell methodology. Biofouling 28:835–842
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this protocol
Cite this protocol
Sternberg, C., Bjarnsholt, T., Shirtliff, M. (2014). Methods for Dynamic Investigations of Surface-Attached In Vitro Bacterial and Fungal Biofilms. In: Donelli, G. (eds) Microbial Biofilms. Methods in Molecular Biology, vol 1147. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-0467-9_1
Download citation
DOI: https://doi.org/10.1007/978-1-4939-0467-9_1
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-0466-2
Online ISBN: 978-1-4939-0467-9
eBook Packages: Springer Protocols