Abstract
The last two decades have witnessed the explosive growth in the development and use of noninvasive neuroimaging technologies that advance the research on human brain under normal and pathological conditions. Multimodal neuroimaging has become a major driver of current neuroimaging research due to the recognition of the clinical benefits of multimodal data, and the better access to hybrid devices. Multimodal neuroimaging computing is very challenging, and requires sophisticated computing to address the variations in spatiotemporal resolution and merge the biophysical/biochemical information. We review the current workflows and methods for multimodal neuroimaging computing, and also demonstrate how to conduct research using the established neuroimaging computing packages and platforms.
Similar content being viewed by others
1 Introduction
Neuroimaging has profoundly advanced neuroscience research and clinical care rapidly in the past two decades, prominently by magnetic resonance imaging (MRI), complemented positron emission tomography (PET), and electroencephalography (EEG)/magnetoencephalography (MEG). The art of neuroimaging today is shaped by three concurrent, interlinked technological developments [1]:
Data Acquisition The advances of imaging instrumentation have enabled digital image acquisition, as well as electronic data storage and communication systems, such as the picture archiving and communication system (PACS). These imaging systems, CT, MRI and PET showed obvious clinical benefits by providing high contrast tissue differentiation. The previous film-based reading was replaced by the electronic displays (axial, coronal and sagittal planes of the volume) without losing diagnostic quality.
Medical Image Computing The growth of neuroimaging has spurred a parallel development of neuroimaging computing methods and workflows, including bias correction, registration, segmentation, information extraction and visualization. We should note the difference between neuroimaging and neuroimaging computing. Neuroimaging focuses on the image acquisition, capturing the snapshot of the brain; whereas neuroimaging computing focuses on the computational analysis of the brain images, extracting and enhancing the information of relevance to best describe the brain anatomy and function.
Package and Platform Development To fit into research and clinical timelines and facilitate translational medicine, the neuroimaging computing methods and workflows are often integrated into software packages. Many such packages were added to imaging systems by the major vendors of medical imaging equipment and many specialized companies. However, a greater number of neuroimaging computing packages and platforms are free and open-source, designed and supported by the medical imaging research groups and communities.
Multimodal neuroimaging, i.e., the simultaneous imaging measurement (EEG/fMRI [2], PET/CT [3]) or summation of separate measurement (PET and sMRI [4], sMRI and dMRI [5], fMRI and dMRI [6]), has become an emerging research area due to better access to imaging devices, especially the hybrid systems, such as PET/CT [7, 8] and PET/MR [9]. The recent advances in neuroimaging computing methods also enabled joint analysis of the multimodal data. The free and open-source software (FOSS) packages and platforms for neuroimaging computing further facilitate the translation of the multimodal neuroimaging research from the lab to better clinical care.
Multimodal neuroimaging advances the neuroscience research by overcoming the limits of individual imaging modalities and by identifying the associations of findings from different imaging sources. Multimodal neuroimaging has been used to investigate a multitude of populations and disorders, such as Alzheimer’s disease (AD) [4, 10–12], schizophrenia [13–16], epilepsy [3, 17–19], obsessive-compulsive disorder (OCD) [20–22], bipolar disorder [23, 24], attention-deficit hyperactivity disorder (ADHD) [25–27], Autism spectrum disorder (ASD) [28–30], traumatic brain injury (TBI) [31–34], stroke [35, 36], multiple sclerosis (MS) [37–39], and brain tumors [9, 40–42]. We have recently reviewed advances in neuroimaging technologies and the applications of multimodal neuroimaging in these neuropsychiatric disorders [43]. Multimodal neuroimaging has also been used in many non-clinical applications, such as building brain machine interface (BMI) [44], tracing neural activity pathways [45] and mapping mind and behavior to brain anatomy [46–48].
Multimodal neuroimaging computing is a very challenging task due to large inter-modality variations in spatiotemporal resolution, and biophysical/biochemical mechanism. Compared to single imaging modality computing, it requires more sophisticated bias correction, co-registration, segmentation, feature extraction, pattern analysis, and visualization. Various methods for neuroimaging analysis have been proposed, and many have been integrated into the task-oriented packages or integrated platforms.
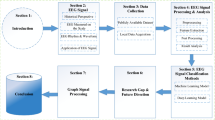
In this paper, we review the state-of-the-art methods and workflows for both modality-specific neuroimaging computing and multimodal neuroimaging computing, and demonstrate how to conduct multimodal neuroimaging research using the established packages and platforms. Fig. 1 provides an overview of the current status and illustrates the major components of neuroimaging computing, including neuroimaging modalities, modality-specific computing workflows (a series of tasks), multimodal computing methods, algorithms, packages, platforms and communities. MRI, PET, EEG/MEG and their computing workflows and methods are discussed in this review. A neuroimaging computing task in an analysis workflow may be fulfilled by multiple algorithms, and the most widely used algorithms, e.g., voxel-based morphometry (VBM) [49], are often integrated into software packages, e.g., Statistical Parametric Mapping (SPM)Footnote 1, FMRIB Software Library (FSL)Footnote 2, and NeurostatFootnote 3. The new imaging tasks also demand the refinement of existing algorithms and development of new algorithms. Similar algorithms are often developed independently in different labs, sometimes with little awareness of existing packages/platforms.
This paper is organized as follows. In Sect. 2, we elaborate the computing workflows, which consist of a number of specific tasks, for individual modalities. In Sect. 3, we review the major multimodal neuroimaging computing methods, i.e., registration, segmentation, feature integration, pattern analysis and visualization. In Sect. 4, we introduce the task-oriented packages and platforms for the tasks mentioned in previous sections. We focus on the free and open source software (FOSS) in this review, since they could help to better realize the quickly evolved methods and workflows than their commercial counterparts, and thus accelerate translational medicine. For the sake of clarity and precision, the algorithms, packages and platforms are not described in detail, but we refer the interested readers to more specific papers instead. In Sect. 5, we give one example of brain tumor surgical planning using the established packages and platforms. Lastly, we outline the future directions of multimodal computing in Sect. 6.
2 Modality-specific neuroimaging computing workflows
2.1 Bias and artifacts correction
Different neuroimaging techniques have different spatiotemporal resolutions, and biophysical/biochemical mechanisms, and thereby require different computing workflows; yet a common step of all workflows is the correction of bias and artifacts in neuroimaging data. The main goal of this task is to remove the data components that contaminate the signals. Tustison recently provided a set of guidelines for managing the instrumental bias when designing and reporting neuroimaging studies [50].
Bias and artifacts in neuroimaging signals may result from imaging systems, environment, and body motion. Many biases and artifacts are induced by the imaging systems, e.g. inhomogeneous radio frequency (RF) coils in MRI, contrast agents in PET/CT, broken or saturated sensors in EEG/MEG system. Environment-related artifacts, arising from generators of magnetic fields outside the human body such as magnetic noise from power lines and other environmental noise sources, such as elevators, air conditioners, nearby traffic, mechanical vibrations transmitted to shielded room, bed vibration, and pulsation [51]. Motion-related artifacts are caused by eye movements, head movements, cardiac and muscular activity, and respiratory motion. The motion of magnetic implements, such as pacemakers and implantable defibrillators [52] may also give rise to artifacts, and may cause danger to patients in strong magnetic field, although there are new MRI compatible pacemakers/defibrillators that have been introduced [53].
The bias and artifacts in MRI are mainly system-related, e.g., RF inhomogeneity causing slice and volume intensity inconsistency. The nonparametric nonuniformity normalization (N3) algorithm and its variant based on Insight Toolkit [54, 55] (N4ITK) [56] are the de facto standard in this area. The acquisition protocols for dMRI are inherently complex, which require fast gradient switching in Echo-Planar Imaging (EPI) and longer scanning time. dMRI is prone to many other types of artifacts, such as eddy current, motion artifacts and gradient-wise inconsistencies [57]. Tortoise [58] and FSL diffusion toolbox (FDT) [59] are popular choices for eddy current correction and motion correction in dMRI data, and the recently proposed DTIPrep [60] offers a thorough solution for all known data quality problems of dMRI. Motion is a serious issue in fMRI, and may lead to voxel displacements in serial fMRI volumes and between slices. Therefore, serial realignment and slice timing correction is required to eliminate the effects of head motion during the scanning session. Linear transformation is usually sufficient for serial alignment, whereas a non-linear auto-regression model is often used for slice timing correction [61]. These two types of correction are commonly performed using SPM and FSL. Dedicated PET scanners have been replaced by the hybrid PET/CT systems [62]. The most commonly seen artifacts on PET/CT are mismatches between CT and PET images caused by body motion due to the long acquisition time of the scan. Metallic implants and contrast agents may also give rise to artifacts on PET/CT, usually leading to overestimation of PET attenuation coefficients and false-positive findings [63]. Knowledge and experience are needed to minimize these artifacts and, in that way, produce better-quality PET/CT images. EEG and MEG signals are often contaminated by all of the three types of artifacts, such as the system-related superconducting quantum interference device (SQUID) jumps, and the noise from the environment or body motion [51]. Visual checks and manual removal are usually required to exclude the artifacts. Another strategy uses signal-processing methods to reduce artifacts while preserving the signal. Linear transformation, e.g., principal component analysis (PCA) and independent component analysis (ICA) [64, 65], and regression, e.g., signal space projection (SSP) and signal space separation (SSS) [66, 67], are frequently applied to the raw EEG/MEG data.
2.2 Structural MRI computing
The sMRI computing workflows usually involve skull striping, tissue and region of interest (ROI) segmentation, surface reconstruction [68], and can include brain morphometry analysis, such as the voxel-based morphometry (VBM)/tensor-based morphometry (TBM)/deformation-based morphometry (DBM) [49], and surface-based morphometry (SBM) [69] by comparing one group of subjects to another or tracking the changes over a sequence of observations for the same subject. FreeSurfer [70] is a well-established tool for brain tissue segmentation and surface reconstruction. When registered into a standard brain space, e.g., the Talariach coordinates [71] and MNI coordinates [72], and labeled with different regions of interest (ROIs) using brain templates, e.g., ICBM template [73] and the AAL template [74], the sMRI datasets can further be analyzed at the ROI level. Various techniques have been investigated to quantitatively analyze the morphological changes in cortex, e.g., grey matter density [49], cortical folding [75], curvedness and shape index [76, 77], cortical thickness [69], and surface area [78, 79], local gyrification index (LGI) [75], and many other shape [78, 80] or texture features [81–83]. Mangin et al. [84] provided an extensive review on the popular morphological features, and Winkler et al. [85] demonstrated how to use these features in imaging genetics.
2.3 Diffusion MRI computing
The dMRI computing workflow consists of four major steps. The first step is to estimate the principle directions of the tensor or the orientation distribution function (ODF) in each voxel, which are used to quantitatively analyze the local white matter morphometry and probe the white matter fiber tracts in the following steps. Advanced fiber orientation estimation methods include the ball and stick mixture models [59], the constrained spherical deconvolution (CSD) [86], the q-ball imaging (QBI) [87], diffusion spectral imaging (DSI) [88], the generalized q-sampling imaging (GQI) [89], and the QBI with Funk Radon and Cosine Transform (FRACT) [90]. Wilkins et al. have provided a detailed comparison of these models [91]. In the second step, various parametric maps based on the tensors/ODFs, i.e., fractional anisotropy (FA), mean diffusivity (MD), radial diffusivity (RD), and axial diffusivity (AXD) maps [92], reveal the focal morphometry of the white matter [93]. The third step is to apply the fiber tracking algorithms [94] to construct 3D models of the white matter tracts, referred to as tractography. Tractography further enables the quantitative analysis of fiber tract morphometry i.e., orientation and dispersion [95], and the analysis of connectome, i.e., connectivity networks of populations of neurons [96]. Brain parcellation and fiber clustering are two major approaches that can separate the neurons into different groups/ROIs, and construct the connectome [97]. Jones et al. [98] recently provided a set of guidelines which define the good practice in dMRI computing.
2.4 Functional MRI computing
After bias and artifacts correction in fMRI, a mean image of the series, or a co-registered anatomical image, e.g., sMRI, is used to estimate some registration coefficients that map it onto a template, followed by spatial smoothing and parameter estimation. Friston [99] gave an introduction to these procedures. When the brain is performing a task, cerebral blood flow (CBF) usually changes as neurons work to complete the task. The primary use of task-evoked fMRI is to identify the correlation between brain activation pattern and brain functions, such as perception, language, memory, emotion and thought [100, 101]. Many models and methods have been suggested to detect patterns of brain activation, and some of them have been integrated into the software packages, such as the general linear model (GLM) in the SPM and FSL packages, and independent component analysis (ICA)/canonical correlation analysis (CCA) in AFNI packageFootnote 4. When brain is at resting state, fMRI is used to detect the spontaneous activation pattern in the absence of an explicit task or stimuli [102]. Resting-state fMRI enables us to deduce the functional connectivity between dispersed brain regions, which form functional brain networks, or resting state networks (RSNs). The Default Mode Network (DMN) is a functional network of several brain regions that show increased activity at rest and decreased activity when performing a task [103]. DMN has been widely used as a measure to compare individual differences in behavior, genetics and neuropathologies, although its use as a biomarker is controversial [104, 105]. Rubinov [106] provided a review of the connectivity measures.
2.5 PET computing
The computing of PET also requires spatial normalization and smoothing, and parameter estimation, similar to fMRI. SPM and Neurostat packages are available for voxel-by-voxel PET analysis. PET functional features are generally pertaining to the radioactive tracers, reflecting particular biochemical process. 2-[18F fluoro-2-deoxy-D-glucose (FDG) is the most widely used tracer to depict glucose metabolism. Several amyloid-binding compounds, \(^{18}\)F-BAY94-9172, \(^{11}\)C-SB-13, \(^{11}\)C-BF-227, \(^{18}\)F-AV-45 and \(^{11}\) C-Pittsburgh compound B (\(^{11}\)C-PiB), have been reported as tracers for imaging amyloid plaques in AD. A number of extensive surveys have been conducted on these amyloid radioactive tracers [107–110]. A variety of static and kinetic parameters can be extracted from the PET data, i.e. the standard uptake value (SUV) [111, 112], cerebral metabolic rate of glucose consumption (CMRGlc) [81, 113], mean index [114], z-scores [115], hypo-metabolic convergence index (HCI) and amyloid convergence index (ACI) [116], tissue time activity curve (TTAC) [117], and difference-of-Gaussian (DoG) parametric maps [118].
2.6 EEG and MEG computing
In EEG and MEG there are usually four components after removing the artifacts or unwanted data components that contaminate the signals. The analysis of event-related potentials (ERP) in EEG or event-related fields (ERF) in MEG aims to analyze brain responses that are evoked by a stimulus or an action, followed by spectral analysis, which transforms the signals into time-frequency domain. The aim of source reconstruction is to localize the neural sources underlying the signals measured at the sensor level. MRI is usually used to provide anatomical reference for source reconstruction. The aim of connectome analysis is to investigate the causality of brain activities and connectivity of brain networks by exploring information flow and interaction between brain regions. Gross et al. provided basic guidelines on EEG and MEG in research [51]. MNEFootnote 5, EEGLABFootnote 6 and eConnectomeFootnote 7 are the most widely used software packages specifically designed for EEG and MEG computing.
3 Multimodal neuroimaging computing methods
3.1 Registration
Registration is the most commonly used technique in a neuroimaging study, and it finds the spatial relationship between two or more images, e.g., multimodal neuroimaging data alignment, serial alignment, and atlas mapping. A registration method can be defined in five aspects, i.e., a cost function for evaluating the similarity between images, a transformation model to determine the degree-of-freedom (DOF) of the deformation, an optimization method for minimizing the cost function, a sampling and interpolation strategy for computation of the cost function, and a multi-resolution scheme for controlling the coarseness of the deformation [119].
Registration methods can be roughly classified into three categories according to the DOF of their transformation models. Rigid registration has a DOF of 6 and allows for global translations and rotations. Affine registration, i.e., linear registration, allows for translation, rotation, scaling and skew of the images. Rigid and affine registration methods are usually sufficient for registering the multimodal datasets of same subject. However, deformable registration, which supports local deformations, is frequently needed to register images with large differences, e.g., registering an image to a template, or registering pre- and post-contrast images of the same subject. Deformable registration always requires rigid or affine registration to obtain a rough initial alignment. In many multimodal studies, a combination of these registration methods were used. For example, we recently jointly analyzed the ADNI FDG-PET and T1-weighted MRI datasets to classify AD and mild cognitive impairment (MCI) patients [79]. The PET images were aligned to MRI using an affine registration method (FSL FLIRT) [120]. The MRI datasets were registered to the MNI template using a deformable method (IRTK) [121], and the output registration coefficients by IRTK were applied to register the PET images to the same template. There are many other widely used registration algorithms, such as B-Spline registration [119, 122], Demons [123], and SyN [124], and ITK [54] registration framework is a standard-bearer for all of these popular registration methods.
3.2 Segmentation
Segmentation is also referred to as brain parcellation or labeling. The brain can be segmented at different levels, i.e., tissues (grey matter, white matter, cerebrospinal fluid), cortical regions, and sub-cortical regions. The segmentation methods can be classified into three categories [125]. The first category is manual and semi-automatic methods, which require manually outlining the brain regions according to a protocol [126, 127] or labeling the landmarks or seed points [128, 129]. These methods are labour-intensive and prone to intra- and inter-operator variation.
The second category is the atlas inverse mapping methods, which can inversely map a labeled atlas, e.g., the standard ICBM and AAL template, or user-defined image, to the original image space. Yao et al. recently provided a review of popular brain atlases [130]. Atlas inverse mapping is simple, but its performance heavily depends on the selected atlas and mapping method.
A more robust but complex solution is multi-atlas labeling, including the multi-atlas propagation with enhanced registration (MAPER) [131] and its variants [132, 133]. These methods carry out whole-brain segmentation in the original image space by fusing multiple labeling results derived from the multiple atlases. Multi-atlas labeling is computationally expensive, but the performance is comparable to manual labeling [125]. FSL FASTFootnote 8 and NifSegFootnote 9 are widely used for brain tissue segmentation. IRTK, Advanced Normalization Tools (ANTs)Footnote 10 and NifRegFootnote 11 are commonly used in multi-atlas labeling as the normalization tools.
3.3 Feature fusion
Various features can be extracted from the neuroimaging data, as described in Sect. 2. Feature fusion is needed to jointly analyze the features from multimodal data. A straightforward solution is to concatenate input multi-view features into a high-dimensional vector, and then apply feature selection methods, such as t-test [134], ANOVA [118], Elastic Net [10, 135], lasso [136] or a combination of these methods [137, 138], to reduce the ’curse of dimensionality’.
These methods show promising results. However, the inter-subject variations cannot be eliminated using the concatenation methods because the inter-subject distances measured by different features may have different scales and variations. With a focus on the subjects, the feature embedding methods, such as multi-view spectral embedding (MSE) [139] and multi-view local linear embedding (MLLE) [140], have been used to explore the geometric structures of local patches in multiple feature spaces and align the local patches in a unified feature space with maximum preservation of the geometric relationships.
In addition, machine learning, especially deep learning, is increasingly used to extract high-level features from neuroimaging data. The advantage of learning-based features is they do not depend on prior knowledge of the disorder or imaging characteristics as the hand-engineered features. They are also essentially suitable for multimodal feature learning, and could expect better performance with larger datasets. However, learning-based features heavily depend on the training datasets [141]. Recently, Suk et al. [142] proposed a feature representation learning framework for multimodal neuroimaging data. One stacked auto-encoder (SAE) was trained for each modality, then the learnt high-level features were further fused with a multi-kernel support vector machine (MK-SVM). They further proposed another deep learning framework based on the deep Boltzmann machine (DBM) and trained it using the 3D patches extracted from the multimodal data [143].
3.4 Pattern analysis
Pattern analysis aims to deduce the patterns of disease pathologies, sensorimotor or cognitive functions in the brain and identify the associated regionally specific effects. A substantial proportion of pattern analysis methods focused on classification of different groups of subjects, e.g., distinguishing AD patients from normal controls [10, 138]. Hinrichs et al. [144, 145] and Zhang et al. [4] recently proposed the multi-kernel support vector machine (MK-SVM) algorithm, which is based on multi-kernel learning and extends the kernel tricks in SVM to the multiple feature spaces. We previously proposed a multifold Bayesian kernelization (MBK) model [79] to transfer the features into diagnosis probability distribution functions (PDFs), and then merge the PDFs instead of the feature spaces.
Regression-based pattern analysis is often used to identify the biomarkers of a group of subjects and probe the boundaries between different groups. The multimodal biomarkers can be based on the voxel features, ROI features and other features, as described in Sect. 2. Regression, such as Softmax regression [10], Elastic Net [135], and lasso [136] can be combined with feature learning in a unified framework.
Recently, the pattern analysis methods have been extended to simulation of future brain development based on the previous states of the brain and comparison to other brains. The basic assumption is that brains with similar cross-sectional and longitudinal deformations would have similar follow-up development [146, 147]. When the population is sufficiently large to include a majority of neurodegenerative changes, the simulated results are more accurate.
3.5 Visualization
The neuroimaging data are mainly 2D and 3D, thus can be visualized in multi-dimensional spaces with 2D and 3D viewers. Multimodal data in 2D space are usually displayed with three layers, including background, foreground and label maps. The 3D viewer enables visualization of volume data, such as volume renderings, triangulated surface models and fiber tracts. Basic image visualization functions, such as look up tables, zoom, window / level, pan, multi-planar reformat, crosshairs, and synchronous pan / scroll for linked viewers, have been implemented in most visualization platforms, such as SlicerFootnote 12 and BioImage SuiteFootnote 13. These platforms also can accommodate visualization of high-dimensional data, e.g., tensors and vector fields.
Image markup refers to the graphical elements overlay, such as fiducials (points), rulers, bounding boxes, and label maps. Image annotation refers to the text-based information [148]. Both image markups and annotations are used to describe the meta information of the images, and annotations can be associated with markup elements as free text.
Another important use of the image markups is interactive visualization. The aforementioned platforms also provide a graphical user interface to interact with the data. For example, the volume rendering module of Slicer allows the users to define a bounding box and visualize the content in the bounding box only. Another module, tractography interactive seeding, is designed for interactive seeding of DTI fiber tracts passing through a list of fiducials or vertices of a 3D model. Slicer also allows the configuration of the layouts and manipulation of content in the viewers to suit a specific use case.
4 FOSS packages and platforms
4.1 Task-oriented packages
The FOSS packages for neuroimaging computing are usually initially designed for a single task, such as registration and segmentation, and some of them then are extended to related tasks and become multifunctional packages. A number of the most widely used FOSS packages are listed in Fig. 1—packages and platforms.
Popular multifunctional packages include FreeSurfer, FSL, SPM, ANTs and NifTK. They cover similar aspects of functionality, but all have particular strengths. FreeSurfer and FSL provide a comprehensive solution of analysis tools for fMRI, sMRI and dMRI data. SPM is designed for the analysis of fMRI, PET, SPECT, EEG and MEG. The recently developed ANTs and NifTK are useful for managing, interpreting and visualizing multimodal data, and represent the state-of-the-art in medical image registration and segmentation. Tustison et al. [149] recently compared ANTs and FreeSurfer in a large-scale evaluation of cortical thickness measurements. Other packages may focus on a specific task or a set of related tasks. IRTKFootnote 14, BRAINs [150], BrainVisaFootnote 15, ITK-SNAPFootnote 16 and MindBoggleFootnote 17 are popular choices for registration and segmentation. In dMRI analysis, CaminoFootnote 18, DTI-TKFootnote 19, DSI StudioFootnote 20, TrackVisFootnote 21 and MRTrixFootnote 22 are most widely used packages. Soares et al. [151] recently conducted a thorough evaluation of these packages and the other dMRI computing packages in published studies. In functional neuroimaging computing, AFNI, PyMVPAFootnote 23 and RESTFootnote 24 are widely used for fMRI analysis, whereas MNE, EEGLAB, eConnectome for EEG/MEG analysis.
4.2 All integrated platforms
For clinical applications, the medical image computing and visualization functions are part of the operation system and must meet the same standards of reliability, robustness, and simplicity of operation as the core imaging equipment. This is usually accomplished using software platforms added onto imaging system by the major vendors of medical image equipment and many specialized companies. Examples include Advantage Windows (General Electric), Syngo Via (Siemens), Vital Image Vitrea (Toshiba), Visage Amira (Visage Imaging), PMOD (PMOD Technologies Ltd.), Definiens (Definiens Inc.), and MimVista (MIM Software Inc.). These packages provide users with a set of analysis tools, compatibility with PACS and customer support. Such clinically oriented systems are not always affordable for academic researchers. Commercial solutions are typically not extensible by the end user, nor oriented towards prototyping of new tools, and may require specialized hardware, thereby limiting their applicability in projects that involve the development of new image computing methods.
As opposed to the commercial platforms, FOSS platforms are meant to provide a research platform that is freely available and does not require specialized hardware. A key step in the evolution of today’s flexible and sophisticated capabilities for image-data-based research medicine was the creation of the 3D Slicer, which is based on a modular architecture [1, 152]. 3D Slicer has become a successful and long-lived platform for the effective use of volumetric images in clinical research and procedure development. There are a number of platforms which aim to cover similar aspects of functionality, e.g., BioImage Suite, BrainSuiteFootnote 25, MIPAVFootnote 26 and MITKFootnote 27.
Some of the libraries contributing to the foundation of Slicer were designed in close collaboration and often share the same developer community. These libraries, including CMake, ITK, VTK and CTK, are distributed as part of the National Alliance for Medical Image Computing (NA-MIC) Kit [153], which are actively supported by the NA-MIC research communityFootnote 28. Many popular packages, e.g., ANTs, MindBoggle, ITK-SNAP, DTIPrep, and MITK are also based on the NA-MIC Kit. NIPYFootnote 29 and NeuroDebianFootnote 30 are another two major research communities for neuroimaging research and platform development. To promote open science, neuroimaging tools and resources are always shared to other community members, usually through the INCFFootnote 31 and NITRCFootnote 32 forums.
Experimental visualization of brain tumor case of DTI Challenge 2015 using 3D Slicer. The panel on the left shows the GUI of the Slicer Mosaic Viewer module previously developed by us. The right side shows the four data viewers, each visualizing a specific step in the surgical planning workflows. The up left viewer shows the registered T1 that overlaid on the DTI volume. The up right viewer shows the segmented tumor (green), ventricle (blue), and motor cortex (red) surfaces. The bottom left viewer shows the reconstructed pial surface of the right hemisphere and white matter surface of the left hemisphere. The bottom right viewer interactively visualizes the peritumoral fiber tracts as the user moves the fiducial. (Color figure online)
5 Example: surgical planning for brain tumor resection
Tractography derived from dMRI has great potential to help neurosurgeons determine tumor resection boundaries in functional areas involving eloquent white matter fibers. The MICCAI DTI ChallengeFootnote 33 is dedicated to comparing different fiber-tracking algorithms in reconstruction of white matter tracts, such as peritumoral tracts and cerebrospinal tract (CST). In this section, we present an example of pre-operative planning for brain tumor resection using the sMRI and dMRI data. The original data consist of a DWI volume and two structure scans of a patient with meningioma. The DWI scan was acquired with a spin-echo EPI sequence with the following parameters: voxel size 2.2 \(\times\) 2.2 \(\times\) 2.2 mm, FOV 220 mm, 58 slices, b-value 1000 s/mm\(^{2}\), 30 diffusion-weighted volumes and 1 baseline volume. The T1 original was acquired using a Ax 3D T1 MPRAGE sequence. The T2 original was acquired using a Ax 3D SPACE sequence.
The original data were computed in four steps, as illustrated in Fig. 2. For dMRI-specific computing, the tensors were estimated using a weighted least square (WLS) algorithm, and the output is a DTI volume. We then registered the T1 and T2 MRI volumes to baseline volume using the affine registration algorithm. The registered T1 and T2 volumes were used as the anatomical references. For sMRI-specific computing, tumor, ventricle and motor cortex were manually seeded and semi-automatically labeled in the baseline volume. The label map of the tumor and ventricle were than used to generate the 3D surface model using the Model Maker module in Slicer. The head surface, pial surface and white matter surfaces for both hemisphere were reconstructed using the Morphologist 2013 pipeline in BrainVisa [68]. For multimodal computing, the white matter tracts were visualized using the Slicer Tractography Interactive Seeding module, which allows users to mark the image with fiducials, and then move it around the tumor to visualize the peritumoral fiber tracts.
6 Future directions
The neuroimaging techniques will keep advancing rapidly, towards higher spatial/temporal/angular resolutions, shorter scanning time, and greater image contrast. In particular, the advances in the hybrid imaging scanners, e.g., PET/CT and PET/MRI, will enter more clinics and laboratories, lowering the cost for data acquisition and enabling more interesting discoveries in a greater multitude of populations and disorders.
The continued growth in the complexity and dimensionality of neuroimaging data will spur the parallel advances of computational models and methods to accommodate such complex data. Such models and methods need to keep increasing the grade of automation, accuracy, reproducibility and robustness, and eventually need to be integrated into the clinical workflows to facilitate clinical testing of the new neuroimaging biomarkers.
The multidisciplinary nature of neuroimaging computing will keep bringing together clinicians, biologists, computer scientists, engineers, physicists, and other researchers who are contributing to, and need to keep abreast of, advances in the neurotechnologies and applications. New methods and models will be developed by the collaboration of different groups or individuals, with rapid iterations. Therefore, future packages and platforms need to respond more quickly to the updates, without compromising the functionality, extensibility and portability. This might cause difficulties in the maintenance of large packages and platforms, but will encourage the researchers to provide smarter solutions, e.g., providing an online version to make the whole process of developing, sharing and updating much quicker for both developers and users.
Notes
References
Kikinis R, Pieper SD, Vosburgh K (2014) 3D Slicer: a platform for subject-specific image analysis, visualization, and clinical support. Intraoper Imaging Image-Guided Therapy 3(19):277–289
He B, Liu Z (2008) Multimodal functional neuroimaging: integrating functional MRI and EEG/MEG. IEEE Rev Biomed Eng 1:23–40
Knopman AA, Wong CH, Stevenson RJ et al (2015) The relationship between neuropsychological functioning and FDG–PET hypometabolism in intractable mesial temporal lobe epilepsy. Epilepsy Behav 44:136–142
Zhang D, Wang Y, Zhou L, Yuan H, Shen D (2011) Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage 55(3):856–867
Savadjiev P, Rathi Y, Bouix S, Smith AR et al (2014) Fusion of white and gray matter geometry: a framework for investigating brain development. Med Image Anal 18:1349–1360
Zhu D, Zhang T, Jiang X, Hu X et al (2014a) Fusing DTI and fMRI data: a survey of methods and applications. NeuroImage 102:184–191
Beyer T, Townsend DW, Brun T, Kinahan PE, Charron M, Robby R et al (2000) A combined PET/CT scanner for clinical oncology. J Nucl Med 41(8):1369–1379
Townsend DW (2001) A combined PET/CT scanner: the choices. J Nucl Med 42(3):533–534
Bisdas S, Nagele T, Schlemmer P, Boss A, Claussen C, Pichler B, Ernemann U (2010) Switching on the lights for real-time multimodality tumor neuroimaging: the integrated positron-emission tomography/MR imaging system. Am J Neuroradiol 31:610–614
Liu SQ, Liu S, Cai W, Che H, Pujol S, Kikinis R, Feng D (2015b) Multi-modal neuroimaging feature learning for multi-class diagnosis of Alzheimer’s disease. IEEE Trans Biomed Eng 62(4):1132–1140
Nir TM, Jahanshad N, Villalon-Reina JE, Toga AW, Jack CR, Weiner MW, Thompson PM (2013) Effectiveness of regional DTI measures in distinguishing Alzheimer’s disease, MCI, and normal aging. NeuroImage 3:180–195
Racine AM, Adluru N, Alexander AL, Christian BT et al. (2014) Associations between white matter microstructure and amyloid burden in precinical Alzheimer’s disease: a multmodal imaging investigation. NeuroImage 4:604–614
Cooper D, Barker V, Radua J, Fusar-Poli P, Lawrie SM (2014) Multimodal voxel-based meta-analysis of structural and functional magnetic resonance imaging studies in those at elevated genetic risk of developing schizophrenia. Psychiatry Res 221(1):69–77
Kochunov P, Chiappelli J, Wright SN, Rowland LM et al (2014) Multimodal white matter imaging to investigate reduced fractional anisotropy and its age-related decline in schizophrenia. Psychiatry Res 223(2):148–156
Liu X, Lai Y, Wang X, Hao C et al (2014b) A combined DTI and structural MRI study in medicated-naive chronic schizophrenia. Magn Reson Imaging 32(1):1–8
Pomarol-Clotet E, Canales-Rodriguez E, Salvador R, Sarro S et al (2010) Medial prefrontal cortex pathology in schizophrenia as revealed by convergent findings from multimodal imaging. Mol Psychiatry 15:823–830
Bonilha L, Keller SS (2015) Quantitative MRI in refractory temporal lobe epilepsy: relationship with surgical outcomes. Quant Imaging Med Surg 5(2):204–224
Fernandez S, Donaire A, Seres E, Setoain X, Bargallo N et al (2015) PET/MRI and PET/MRI/SISCOM coregistration in the presurgical evaluation of refractory facol epilepsy. Epilepsy Res 111:1–9
Abela E, Rummel C, Hauf M, Weisstanner C, Schindler K, Wiest R (2014) Neuroimaging of epilepsy: lesions, networks, oscillations. Clin Neuroradiol 24(1):5–15
Agam Y, Vangel M, Roffman JL, Gallagher PJ et al (2014) Dissociable genetic contributions to error processing: a multimodal neuroimaging study. PLoS ONE 9(7):e101,784
Radua J, Grau M, van den Heuvel OA, de Schotten MT et al (2014) Multimodal voxel-based meta-analysis of white matter abnormalities in obsessive-compulsive disorder. Neuropsychopharmacology 39(7):1547–1557
Taylor SF, Stern ER, Gehring WJ (2007) Neural systems for error monitoring: recent findings and theoretical perspectives. Neuroscientist 13(2):160–172
Phillips ML, Swartz HA (2014) A critical appraisal of neuroimaging studies of bipolar disorder: toward a new conceptualization of underlying neural circuitry and a road map for future research. Am J Psychiatry 171(8):829–843
Sui J, Pearlson GD, Caprihan A, Adali T, Kiehl KA et al (2011) Discriminating schizophrenia and bipolar disorder by fusing fMRI and DTI in a multimodal CCA+ joint ICA model. NeuroImage 57(3):839–855
Anderson A, Douglas PK, Kerr WT, Haynes VS et al (2014) Non-negative matrix factorization of multimodal MRI, fMRI and phenotypic data reveals differential changes in default mode subnetworks in ADHD. NeuroImage 102(1):207–219
Dai D, Wang J, Hua J, He H (2012) Classification of ADHD children through multimodal magnetic resonance imaging. Front Syst Neurosci 6(63):1–8
Hong SB, Zalesky A, Fornito A, Park S, Yang YH et al (2014) Connectomic disturbances in attention-deficit/hyperactivity disorder: a whole-brain tractography analysis. Biol Psychiatry 76(8):656–663
Anagnostou E, Taylor MJ (2011) Review of neuroimaging in Autism spectrum disorders: what have we learnt and where we go from here. Mol Autism 2(4):1–9
Mueller S, Keeser D, Samson AC, Kirsch V, Blautzik J et al. (2013) Convergent Findings of Altered Functional and Structural Briain Connectivity in Individuals with High Functioning Autism: A Multimodal MRI Study. PLoS ONE 8(6):e67,329
Stigler KA, McDonald BC, Anand A et al (2011) Structural and functional magnetic resonance imaging of Autism spectrum disorders. Brain Res 1380:146–161
Cherubini A, Luccichenti G, Peran P, Hagberg GE et al (2007) Multimodal fMRI tractography in normal subjects and in clinically recovered traumatic brain injury patients. NeuroImage 34(4):1331–1341
Dean PJ, Sato JR, Vieira G, McNamara A, Sterr A (2014) Multimodal imaging of mild traumatic brain injury and persistent postconcussion syndrome. Brain Behav 5(1):45–61
Irimia A, Chambers MC, Alger JR, Filippou M, Prastawa MW et al (2011) Comparison of acute and chronic traumatic brain injury using semi-automatic multimodal segmentation of MR volumes. J Neurotrauma 28(11):2287–2306
Turken AU, Herron TJ, Kang X, O’Connor LE, Sorenson DJ et al. (2009) Multimodal surface-based morphometry reveals diffuse cortical atrophy in traumatic brain injury. BMC Medical Imaging 9(20)
Copen WA (2015) Multimodal imaging in acute ischemic stroke. Curr Treat Options Cardiovas Med 17(10):1–17
Tong E, Hou Q, Fiebach JB, Wintermark M (2014) The role of imaging in acute ischemic stroke. Neurosurg Focus 36(1):E3
Achiron A, Barak Y (2003) Cognitive impairment in probable multiple sclerosis. J Neurol Neurosurg Psychiatry 74:443–446
Louapre C, Perlbarg V, Garcia-Lorenzo D, Urbanski M et al (2014)Brain networks disconnection in early multiple sclerosis cognitive deficits: an anatomofunctional study. Hum Brain Map 35:4706–4717
Tona F, Petsas N, Sbardella E, Prosperini L et al (2014) Multiple sclerosis: altered thalamic resting-state functional connectivity and its effect on cognitive function. Radiology 271(3):814–821
Durst CR, Raghavan P, Shaffrey ME, Schiff D et al (2014) Multimodal MR imaging model to predict tumor infiltration in patients with gliomas. Neuroradiology 56(2):107–115
Neuner I, Kaffanke JB, Langen KJ, Kops ER, Tellmann L et al (2012) Multimodal imaging utilising integrated MR-PET for human brain tumor assessment. Eur Radiol 22:2568–2580
Tempany CM, Jayender J, Kapur T, Bueno R et al (2014) Multimodal imaging for improved diagnosis and treatment of cancers. Cancer 121(6):817–827
Liu S, Cai W, Liu SQ, Zhang F, Fulham M, Feng D, Pujol S, Kikinis R (2015a) Multimodal neuroimaging computing: a review of the applications in neuropsychiatric disorders. Brain Info 2(3). doi:10.1007/s40708-015-0019-x
Morioka H, Kanemura A, Morimoto S, Yoshioka T et al (2013) Decoding spatial attention by using cortical currents estimated from electroencephalography with near-infrared spectroscopy prior information. NeuroImage 90:128–139
Liu Z, Ding L, He B (2006) Integration of EEG/MEG with MRI and fMRI in functional neuroimaging. IEEE Eng Med Biol Mag 25(4):46–53
Binder JR, Desai RH, Graves WW, Conant LL (2009) Where is the semantic system? A critical review and meta-analysis of 120 functional neuroimaging studies. Cereb Cortex 19(12):2767–2796
Nguyen VT, Cunnington R (2014) The superior temporal sulcus and the N170 during face processing: single trial analysis of concurrent EEG–fMRI. NeuroImage 86:492–502
Okamoto M, Dan K, Shimizu K, Takeo K et al (2004) Multimodal assessment of cortical activation during apple peeling by NIRS and fMRI. NeuroImage 21(4):1275–1288
Ashburner J, Friston JK (2000) Voxel-based morphometry: the methods. NeuroImage 11(6):805–821
Tustison NJ, Johnson HJ, Rohlfing T, Klein A et al. (2013) Instrumentation bias in the use and evaluation of scientific software: recommendations for reproducible practices in the computational sciences. Front Neurosci 7(162)
Gross J, Baillet S, Barnes GR, Henson RN, Hillebrand A et al (2013) Good practice for conducting and reporting MEG research. NeuroImage 65:349–363
Gotte M, Russel I, de Roest G, Germans T, Veldkamp R et al (2010) Magnetic resonance imaging, pacemakers and implantable cardioverter-defibrillators: current situation and clinical perspective. Neth Heart J 18(1):31–37
Bovenschulte H, Schluter-Brust K, Liebig T, Erdmann E, Eysel P, Zobel C (2012) MRI in patients with pacemakers: overview and procedural management. Deutsch Arztebl Int 109(15):270–275
Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC (2014) The insight toolkit image registration framework. Front Neuroinf 8(1):1–13
Yoo TS, Ackerman MJ, Lorensen WE, Schroeder W et al (2002) Engineering and algorithm design for an image processing API: a technical report on ITK: the Insight Toolkit. Stud Health Technol Inf 85:586–592
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29(6):1310–1320
Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE (2012) Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage 59(3):2142–2154
Pierpaoli C, Walker L, Irfanoglu M et al. (2010) TORTOISE: an integrated software package for processing of diffusion MRI data. In: The 18th ISMRM Annual Meeting, vol 1597
Behrens T, Woolrich M, Jenkinson M, Johansen-Berg H et al (2003) Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med 50(5):1077–1088
Oguz I, Farzinfar M, Matsui J, Budin F, Liu Z, Gerig G et al (2014) DTIPrep: quality control of diffusion-weighted images. Front Neuroinf 8(4):1–11
Friston KJ, Williams S, Howard R, Frackwiak RSJ, Tumer R (1996) Movement-related effects in fMRI time-series. Magn Reson Med 35(3):346–355
Hricak H, Choi BI, Scott AM, Sugimura K et al (2010) Global trends in hybrid imaging. Radiology 257(2):498–506
Sureshbabu W, Mawlawi O (2005) PET/CT imaging artifacts. J Nucl Med Technol 33(3):156–161
Gross J, Ioannides A (1999) Linear transforms of data space in MEG. Phys Med Biol 44:2081–2097
Vigario R (1997) Extraction of ocular artifacts from EEG using independent component analysis. Electroencephalogr Clin Neurophysiol 103:295–404
Taulu S, Simola J (2006)Spatiotemporal signal separation method for rejecting nearby interference in MEG measurements. Phys Med Biol 51:1759–1768
Uusitalo M, Ilmoniemi R (1997) Signal-space projection method for separating MEG or EET into components. Med Biol Eng Comput 35:135–140
Geffroy D, Rivière D, Denghien I, Souedet N, Laguitton S, Cointepas Y (2011) BrainVISA: a complete software platform for neuroimaging. In: Python in neuroscience workshop
Fischl B, Dale AM (2000) Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci 97(20):11,050–11,055
Dale AM, Fischl B, Sereno MI (1999) Cortical surface-based analysis: I segmentation and surface reconstruction. NeuroImage 9(2):179–194
Talariach J, Tournoux P (1988) Co-planar stereotaxic atlas of the human brain 3-dimentional proportional system: an approach to cerebral imaging. Stuttgart, New York
Fonov V, Evans A, Botteron K, Almli C et al (2010) Unbiased average age-approapriate atlases for pediatric studies. NeuroImage 54(1):313–327
Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K et al (2001) A probabilistic atlas and reference system for the human brain: international consortium for brain mapping (ICBM). Philos Trans R Soc London Ser 356(1412):1293–1322
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F et al (2002) Automated anatomical labelling of activations in SPM using a macroscopy anatomical pacellation of the MNI MRI single-subject brain. NeuroImage 15(1):273–289
Schaer M, Cuadra M, Tamarit L, Lazeyras F, Eliez S, Thiran J (2008) AA surface-based approach to quantify local cortical gyrification. IEEE Trans Med Imaging 27(2):161–170
Awate SP, Yushkevich PA, Song Z, Licht DJ, Gee JC (2010) Cerebral cortical folding analysis with multivariate modeling and testing: studies on gender differences and neonatal development. NeuroImage 53(2):450–459
Cash DM, Melbourne A, Modat M, Cardoso MJ, Clarkson MJ, Fox NC, Ourselin S (2012) Cortical folding analysis on patients with Alzheimer’s disease and mild cognitive impairment. In: Ayache N, Delingette H, Golland P, Mori K (eds) Medical image computing and computer-assisted intervention (MICCAI), LNCS, vol 7512. Springer, Berlin, pp 289–296
Liu S, Cai W, Song Y, Pujol S, Kikinis R, Wen L, Feng D (2013a) Localized sparse code gradient in Alzheimer’s disease staging. In: The 35th annual international conference of the IEEE engineering in medicine and biology society (EMBC), IEEE, pp 5398–5401
Liu S, Song Y, Cai W, Pujol S, Kikinis R, Wang X, Feng D (2013c) Multifold Bayesian kernelization in Alzheimer’s diagnosis. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N (eds) The 16th international conference on medical image computing and computer-assisted intervention (MICCAI), LNCS, vol 8150. Springer, Berlin Heidelberg, pp 303–310
Liu S, Zhang L, Cai W, Song Y, Wang Z, Wen L, Feng D (2013d) A supervised multiview spectral embedding method for neuroimaging classification. In: The 20th IEEE international conference on image processing (ICIP), pp 601–605
Cai W, Liu S, Wen L, Eberl S, Fulham MJ, Feng D (2010) 3D neurological image retrieval with localized pathology-centric CMRGlc patterns. In: The 17th IEEE international conference on image processing (ICIP), pp 3201–3204
Liu S, Cai W, Wen L, Eberl S, Fulham M, Feng D (2011a) Localized functional neuroimaging retrieval using 3D discrete curvelet transform. In: IEEE international symposium on biomedical imaging: from nano to macro (ISBI), pp 1877–1880
Liu S, Cai W, Wen L, Feng D (2012) Multiscale and multiorientation feature extraction with degenerative patterns for 3D neuroimaging retrieval. In: The 19th IEEE international conference on image processing (ICIP), pp 1249–1252
Mangin J, Jouvent E, Cachia A (2010) In-vivo measurement of cortical morphology: means and meanings. Curr Opin Neurol 23(4):359–367
Winkler AM, Kochunov P, Blangero J et al (2010) Cortical thickness or grey matter volume? the importance of selecting the phenotype for imaging genetics studies. NeuroImage 53(3):1135–1146
Tournier JD, Calamante F, Connelly A (2007) Robust determination of the fiber orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. NeuroImage 35(4):1459–1472
Tuch DS (2004) Q-ball imaging. Magn Reson Med 52:1358–1372
Wedeen V, Hagmann P, Tseng W, Reese T, Weisskoff R (2005) Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn Reson Med 54(6):1377–1386
Yeh F, Wedeen V, Tseng W (2010) Generalized Q-sampling imaging (GQI). IEEE Trans Med Imaging 29:1626–1635
Haldar JP, Leahy RM (2013) Linear transforms for Fourier data on the sphere: application to high angular resolution diffusion MRI of the brain. NeuroImage 71:233–247
Wilkins B, Lee N, Gajawelli N, Law M, Lepore N (2015) Fiber estimation and tractography in diffusion MRI: development of simulated brain images and comparison of multi-fiber analysis methods at clinical b-values. NeuroImage 109:341–356
Maier-Hein KH, Westin CF, Shenton ME, Weiner MW, Raj A, Thomann P et al (2014) Widespread white matter degeneration preceding the onset of dementia. Alzheimer’s Dementia S1552–5260(14):1–9
Savadjiev P, Kindlemann G, Bouix S, Sheton M, Westin C (2010) Local white matter geometry from diffusion tensor gradients. NeuroImage 49:3175–3186
Mori S, van Ziji PC (2002) Fiber tracking: principles and strategies: a technical review. NMR Biomed 15(7–8):468–480
Durrieman S, Pennec X, Trouve A, Ayache N (2009) Statistical models of sets of curves and surfaces based on currents. Med Image Anal 13(5):793–808
Zalesky A, Fornito A, Harding IH et al (2010) Whole-brain anatomical networks: does the choice of nodes matter? NeuroImage 50(3):970–983
O’Donnell LJ, Golby AJ, Westin CF (2013) Fiber clustering versus the parcellation-based connectome. NeuroImage 80:283–289
Jones DK, Knosche TR, Turner R (2013) White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion MRI. NeuroImage 73:239–254
Friston KJ (2003) Introduction: experimental design and statistical parametric mapping. In: Frackowiak RS et al (eds) Human brain function, 2nd edn. Elsevier, New York
Davison EN, Schlesinger KJ, Bassett DS, Lynall MR et al (2015) Brain network adaptability across task states. PLoS Comput Biol 11(1):e1004,029
Turk-Browne NB (2013) Functional interactions as big data in the human brain. Science 342(6158):580–584
Biswal B, Yetkin F, Haughton V, Hyde J (1995) Functional connectivity in motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 34:537–541
Raichle M, MacLeod A, Snyder A, Powers W et al (2001) A default mode of brain function. Proc Natl Acad Sci 98(2):676–682
Buckner R, Andrews-Hanna J, Schacter D (2008) The brain’s default network: anatomy, function and relevance to disease. Ann N Y Acad Sci 1124:1–38
Jiang T, Liu Y, Shi F, Shu N, Liu B et al (2008) Multimodal magnetic resonance imaging for brain disorders: advances and perspectives. Brain Imaging Behav 2:249–257
Rubinov M, Sporns O (2010) Complex network measures of brain connectivity: uses and interpretations. NeuroImage 52:1059–1069
Carpenter AJ, Pontecorvo M, Hefti F, Skovronsky D (2009) The use of the exploratory IND in the evaluation and development of 18F-PET radiopharmaceuticals for amyloid imaging in the brain: a review of one company’s experience. Q J Nucl Med Mol Imaging 53(4):387–393
Ni R, Gillberg P, Bergfors A, Marutle A, Nordberg A (2013) Amyloid tracers detect multiple binding sites in Alzheimer’s disease brain tissue. Brain 136(7):2217–2227
Perrin RJ, Fagan AM, Holtzmann DM (2009) Multimodal techniques for diagnosis and prognosis of Alzheimer’s disease. Nature 461:916–922
Thompson PM, Ye L, Morgenstem JL, Sue L, Beach TG et al (2009) Interaction of the amyloid imaging tracer FDDNP with hallmark Alzheimer’s disease pathologies. J Neurochem 109(2):623–630
Clark CM, Pontecorvo MJ, Beach TG, Bedell BJ, Coleman RE, Doraiswamy PM et al (2012) Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid- plaques: a prospective cohort study. Lancet Neurol 11(8):669–678
Landau SM, Lu M, Joshi AD, Pontecorvo M, Mintun MA, Trojanowski JQ, Shaw LM, Jagust WJ, Initiative Alzheimer’s Disease Neuroimaging (2013) Comparing positron emission tomography imaging and cerebrospinal fluid measurements of beta-amyloid. Ann Neurol 74(6):826–836
Sokoloff L, Reivich M, Kennedy C, Des-Rosiers M et al (1977) The [14C]deoxy-glucose method for the measurement of local cerebral glucose utilization: theory, procedure and normal values in the consicious and anesthetized albino Rat. J Neurochem 28:897–916
Batty S, Clark J, Fryer T, Gao X (2008) Prototype system for semantic retrieval of neurological PET images. In: Gao X, Müller H, Loomes M, Comley R, Luo S (eds) Medical imaging and informatics, LNCS, vol 4987. Springer, Berlin Heidelberg, pp 179–188
Minoshima S, Frey KA, Koeppe RA, Foster NL, Kuhl DE (1995) A diagnostic approach in Alzheimer’s disease using three-dimensional stereotactic surface projections of fluorine-18-FDG PET. J Nucl Med 36(7):1238–1248
Chen K, Ayutyanont N, Langbaum JB, Fleisher AS, Reschke C et al (2011) Characterizing Alzheimer’s disease using a hypometabolic convergence index. NeuroImage 56(1):52–60
Cai W, Feng D, Fulton R (2000) Content-based retrieval of dynamic PET functional images. IEEE Trans Inf Technol Biomed 4(2):152–158
Cai W, Liu S, Song Y, Pujol S, Kikinis R, Feng D (2014) A 3D difference of Gaussian based lesion detector for brain PET. In: IEEE international symposium on biomedical imaging: from nano to macro (ISBI), pp 677–680
Klein S, Staring M, Murphy K, Viergever MA, Pluim JP (2010) elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29(1):196–205
Jenkinson M, Bannister P, Brady M, Smith S (2002) Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage 17(2):825–841
Rueckert D, Sonoda L, Hayes C et al (1999) Non-rigid registration using free-form deformations: applications to breast MR images. IEEE Trans Med Imaging 18(8):712–721
Murphy K, van Ginneken B, Reinhardt J, Kabus S et al (2011) Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge. IEEE Trans Med Imaging 30:1901–1920
Vercauteren T, Pennec X, Perchant A, Ayache N (2009) Diffeomorphic demons: efficient non-parametric image registration. NeuroImage 45(Suppl. 1):S61–S72
Avants B, Epstein C, Grossman M, Gee J (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12(1):26–41
Heckemann RA, Keihaninejad S, Aljabar P, Gray KR, Nielsen C, Rueckert D et al (2011) Automatic morphometry in Alzheimer’s disease and mild cognitive impairment. NeuroImage 56(4):2024–2037
Hammers A, Allom R, Koepp M et al (2003) Three-dimentional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Map 19(4):224–247
Shattuck D, Mirza M, Adisetiyo V et al (2008) Construction of a 3D probabilistic atlas of human cortical structures. NeuroImage 39(3):1064–1080
Csernansky J, Wang L, Joshi S et al (2004) Computational anatomy and neuropsychiatric disease: probabilistic assessment of variation and statistical inference of group difference, hemispheric asymmetry and time-dependent change. NeuroImage Suppl 1(23):56–68
Rademacher J, Galaburda A, Kennedy D et al (1992)Human cerebral cortex: localization, parcellation, and morphometry with magnetic resonance imaging. J Cogn Neurosci 4(4):352–374
Yao Z, Hu B, Xie Y, Moore P, Zheng J (2015) A review of structural and functional brain networks: small world and atlas. Brain Inf 2(1):45–52
Heckemann R, Hajnal J, Aljabar P, Rueckert D, Hammers A (2006) Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage 33(1):115–126
Aljabar P, Heckemann R, Hammers A et al (2009) Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. NeuroImage 46:726–738
Heckemann R, Keihaninejad S, Aljabar P, Rueckert D et al (2010) Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. NeuroImage 51(1):221–227
Liu S, Cai W, Wen L, Eberl S, Fulham MJ, Feng D (2011b) Generalized regional disorder-sensitive-weighting scheme for 3D neuroimaging retrieval. In: The 33rd annual international conference of the IEEE engineering in medicine and biology society (EMBC), pp 7009–7012
Shen L, Kim S, Qi Y, Inlow M, Swaminathan S, Nho K, Wan J, Risacher S, Shaw L, Trojanowski J, Weiner M, Saykin A (2011) Identifying neuroimaging and proteomic biomarkers for MCI and AD via the elastic net. In: Liu T, Shen D, Ibanez L, Tao X (eds) Multimodal brain image analysis (MBIA), LNCS, vol 7012. Springer, Berlin Heidelberg, pp 27–34
Zhu X, Suk HI, Shen D (2014b) A novel matrix-similarity based loss function for joint regression and classification in AD diagnosis. NeuroImage 100:91–105
Liu S, Cai W, Wen L, Feng D (2013b) Multi-channel brain atrophy pattern analysis in neuroimaging retrieval. In: IEEE international symposium on biomedical imaging: from nano to macro (ISBI), pp 206–209
Liu S, Cai W, Wen L, Feng DD, Pujol S, Kikinis R, Fulham MJ, Eberl S (2014a) Multi-channel neurodegenerative pattern analysis and its application in Alzheimer’s disease characterization. Comput Med Imaging Graph 38:436–444
Xia T, Tao D, Mei T, Zhang Y (2010) Multiview spectral embedding. EEE Trans Syst Man Cybern Part B 40(6):1438–1446
Shen H, Tao D, Ma D (2013) Multiview locally linear embedding for effective medical image retrieval. PLoS ONE 8(12):e82,409
Bengio Y, Courville A, Vincent P (2013) Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 35(8):1798–1828
Suk HI, Lee S, Shen D (2013) Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Struct Funct 220(2):841–959
Suk H, Lee S, Shen D (2014) Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. NeuroImage 101:569–582
Hinrichs C, Singh V, Xu G, Johnson S (2009) MKL for robust multi-modality AD classification. In: Yang G (ed) Medical image computing and computer-assisted intervention (MICCAI), LNCS, vol 5762. Springer, New York, pp 786–794
Hinrichs C, Singh V, Xu G, Johnson S (2011) Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. NeuroImage 55:574–589
Liu SQ, Liu S, Zhang F, Cai W, Pujol S, Kikinis R, Feng D (2015c) Longitudinal brain MR retrieval with diffeomorphic demons registration: what happened to those patients with similar changes? In: IEEE international symposium on biomedical imaging: from nano to macro (ISBI), IEEE
Modat M, Simpson I, Cardoso M, Cash D et al. (2014) Simulating neurodegeneration through longitudinal population analysis of structural and diffusion weighted MRI data. Medical image computing and computer-assisted intervention (MICCAI), LNCS, vol 8675. Springer, Berlin, pp 57–64
Rubin D, Mongkolwat P, Kleper V et al (2009) Annotation and image markup: accessing and interoperating with the semantic content in medical imaging. IEEE Intell Syst 24(1):57–65
Tustison NJ, Cook PA, Klein A, Song G et al (2014) Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. NeuroImage 99:166–179
Pierson R, Johnson HJ, Harris G, Keefe H et al (2011) Fully automated analysis using BRAINS: AutoWorkup. NeuroImage 54:328–336
Soares JM, Marques P, Alves V, Sousa N (2013) A hitchhiker’s guide to diffusion tensor imaging. Front Neurosci 7:31
Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C et al (2012) 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30(9):1323–1341
Pieper S, Lorensen B, Schroeder W, Kikinis R (2006) The NA-MIC Kit: ITK, VTK, pipelines, grids and 3D Slicer as an open platform for the medical image computing community. In: The 3rd IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), IEEE, pp 698–701
Acknowledgments
This work was supported by supported by ARC, NA-MIC (NIH U54EB005149), and NAC (NIH P41RR013218).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Liu, S., Cai, W., Liu, S. et al. Multimodal neuroimaging computing: the workflows, methods, and platforms. Brain Inf. 2, 181–195 (2015). https://doi.org/10.1007/s40708-015-0020-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40708-015-0020-4