Abstract
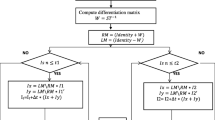
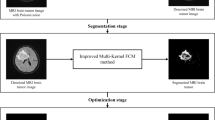
Brain is most complex and central part of the human body. Millions of cells are present in the brain. Brain tumor is extra unwanted cell in the human brain. It is mainly categorized into benign and malignant. Benign tumor cells have very similar characteristics with its surrounding cells. Its accurate detection and segmentation is very challenging task. Image segmentation methods have major contribution in detection and segmentation of these tumor cells. Segmentation methods are either boundary based or region based. These methods use traditional integral-order calculus. It has been observed that these approaches are unable to detect low variational region such as benign tumor. In the present manuscript, fractional diffusion-based benign brain tumor detection and segmentation method is being proposed. It has been observed that the proposed method is able to detect and segment benign brain tumor region more accurately. Higher accuracy has been obtained due to fractional-order derivative. Frequency domain derivative definition has been used in the proposed method due to simplicity and low computational cost. A hardware model of the proposed work has been also presented in the current manuscript. The results obtained have been compared with existing boundary-based and region-based tumor detection and segmentation methods. It has been found that the proposed method is having higher accuracy in benign brain tumor detection and segmentation with low computational cost.





Similar content being viewed by others
References
R.S. Fager, K.V. Peddanarappagari, G.N. Kumar, Pixel-based reconstruction (PBR) promising simultaneous techniques for CT reconstructions. IEEE Trans. Med. Imaging MI 12(1), 4–9 (1993). https://doi.org/10.1109/42.222660
American Brain Tumor Association. http://www.abta.org/
M. Bajpai, P. Munshi, P. Gupta, C. Schorr, M. Maisl, High resolution 3D image reconstruction using algebraic method for X-ray cone-beam geometry over circular and helical trajectories. NDT & E Int. 60, 62–69 (2013). https://doi.org/10.1016/j.ndteint.2013.07.009
B. Mughal, M. Sharif, N. Muhammad, Bi-model processing for early detection of breast tumor in CAD system. Eur. Phys. J. Plus 132(6), 266 (2017). https://doi.org/10.1140/epjp/i2017-11523-8
B. Mughal, N. Muhammad, M. Sharif, Deviation analysis for texture segmentation of breast lesions in mammographic images. Eur. Phys. J. Plus 133(11), 455 (2018). https://doi.org/10.1140/epjp/i2018-12294-4
R.C. Gonzalez, R.E. Woods, Digital Image Processing, 3rd edn. (Pearson, London, 2014)
D. Terzopoulos, K. Fleischer, Deformable models. Vis. Comput. 4, 306–331 (1988)
A.M. Hasan, F. Meziane, R. Aspin, H.A. Jalab, Segmentation of brain tumors in MRI images using three-dimensional active contour without edge. Symmetry (2016). https://doi.org/10.3390/sym8110132
M. Kassa, A. Witkin, D. Terzopoulos, Snakes: active contour models. Int. J. Comput. Vis. 4, 321–331 (1987)
A. Islam, S.M.S. Reza, K.M. Iftekharuddin, Multifractal texture estimation for detection and segmentation of brain tumors. IEEE Trans. Biomed. Eng. 60(11), 3204–3215 (2013). https://doi.org/10.1109/TBME.2013.2271383
X. Ren, J. Malik, Learning a classification model for segmentation. In: Proceedings Ninth IEEE International Conference on Computer Vision, vol. 1, pp. 10–17 (2003). https://doi.org/10.1109/ICCV.2003.1238308
W. Pieczynski, Statistical image segmentation. Mach. Graph. Vis. 1, 261–268 (1992)
S.W. Zucker, R.A. Hummel, A three-dimensional edge operator. IEEE Trans. Pattern Anal. Mach. Intell. PAMI 3(3), 321–324 (1981)
D.G. Morgenthaler, A. Rosenfeld, Multidimensional edge detection by hypersurface fitting. IEEE Trans. Pattern Anal. Mach. Intell. PAMI 3(4), 482–486 (1981). https://doi.org/10.1109/TPAMI.1981.4767134
W. Deng, W. Xiao, H. Deng, J. Liu, MRI brain tumor segmentation with region growing method based on the gradients and variances along and inside of the boundary curve. In: 2010 3rd International Conference on Biomedical Engineering and Informatics, vol. 1, pp. 393–396 (2010). https://doi.org/10.1109/BMEI.2010.5639536
S.Z. Oo, A.S. Khaing, Brain tumor detection and segmentation using watershed segmentation and morphological operation Int. J. Res. Eng. Technol. 3(3), 367–374 (2014)
E. Abdel-Maksoud, M. Elmogy, R. Al-Awadi, Brain tumor segmentation based on a hybrid clustering technique. Egypt. Inform. J. 16(1), 71–81 (2015). https://doi.org/10.1016/j.eij.2015.01.003
B. Al-Naami, A. Bashir, H. Amasha, J. Al-Nabulsi, A.M. Almalty, Statistical approach for brain cancer classification using a region growing threshold. J. Med. Syst. 35(4), 463–471 (2011). https://doi.org/10.1007/s10916-009-9382-6
K.B. Oldham, J. Spanier, “The Fractional Calulus” Theory and Applications of Differentiation and Integration of Arbitrary Order, 1st edn. (Academic Press, New York, 2006)
S.K. Chandra, M. Kumar Bajpai, Effective algorithm for benign brain tumor detection using fractional calculus. In: TENCON 2018–2018 IEEE Region 10 Conference, 2018, pp. 2408–2413. https://doi.org/10.1109/TENCON.2018.8650163
T. Wei, Y. Li, Identifying a diffusion coefficient in a time-fractional diffusion equation. Math. Comput. Simul. 151, 77–95 (2018). https://doi.org/10.1016/j.matcom.2018.03.006
J. Bai, X.C. Feng, Fractional-order anisotropic diffusion for image denoising. IEEE Trans. Image Process. IP 16(10), 2492–2502 (2007). https://doi.org/10.1109/TIP.2007.904971
J.J. Koenderink, The structure of images. Biol. Cybern. 50(5), 363–370 (1984)
S.K. Chandra, M. Kumar Bajpai, Fractional anisotropic diffusion for image denoising. In: 2018 IEEE 8th International Advance Computing Conference (IACC), pp. 344–348 (2018). https://doi.org/10.1109/IADCC.2018.8692094
C.B. Gao, J.L. Zhou, J.R. Hu, F.N. Lang, Edge detection of colour image based on quaternion fractional differential. IET Image Process. 5(3), 261–272 (2011). https://doi.org/10.1049/iet-ipr.2009.0409
M. Polak, H. Zhang, M. Pi, An evaluation metric for image segmentation of multiple objects. Image Vis. Comput. 27(8), 1223–1227 (2009). https://doi.org/10.1016/j.imavis.2008.09.008
C. Zhao, W. Shi, Y. Deng, A new Hausdorff distance for image matching. Pattern Recogn. Lett. 26(5), 581–586 (2005). https://doi.org/10.1016/j.patrec.2004.09.022
S.K. Chandra, M.K. Bajpai, Mesh free alternate directional implicit method based three dimensional super-diffusive model for benign brain tumor segmentation. Comput. Math. Appl. (2019). https://doi.org/10.1016/j.camwa.2019.02.009
B.H. Menze et al., The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging (2014). https://doi.org/10.1109/TMI.2014.2377694
Acknowledgements
The authors would like to thank Dr. Pritee Khanna, Convener, Computer Vision and Image Processing Lab, Indian Institute of Information Technology, Design & Manufacturing Jabalpur, for supplying computational support to carry out experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Chandra, S.K., Bajpai, M.K. Efficient three-dimensional super-diffusive model for benign brain tumor segmentation. Eur. Phys. J. Plus 135, 419 (2020). https://doi.org/10.1140/epjp/s13360-020-00414-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1140/epjp/s13360-020-00414-8