Abstract
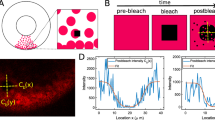
Fluorescence recovery after photobleaching (FRAP) is an experimental technique used to measure the mobility of proteins within the cell nucleus. After proteins of interest are fluorescently tagged for their visualization and monitoring, a small region of the nucleus is photobleached. The experimental FRAP data are obtained by recording the recovery of the fluorescence in this region over time.
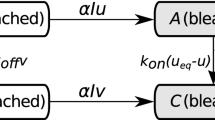
In this paper, we characterize the fluorescence recovery curves for diffusing nuclear proteins undergoing binding events with an approximate spatially homogeneous structure. We analyze two mathematical models for interpreting the experimental FRAP data, namely a reaction-diffusionmodel and a compartmental model. Perturbation analysis leads to a clear explanation of two important limiting dynamical types of behavior exhibited by experimental recovery curves, namely, (1) a reduced diffusive recovery, and (2) a biphasic recovery characterized by a fast phase and a slow phase. We show how the two models, describing the same type of dynamics using different approaches, relate and share common ground. The results can be used to interpret experimental FRAP data in terms of protein dynamics and to simplify the task of parameter estimation. Application of the results is demonstrated for nuclear actin and type H1 histone.
Similar content being viewed by others
References
Axelrod, D., D. E. Koppel, J. Schlessinger, E. Elson and W. W. Webb (1976). Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16, 1055–1069.
Bender, C. M. and S. A. Orszag (1978). Advanced Mathematical Methods for Scientists and Engineers, New York: McGraw-Hill.
Berg, H. C. (1993). Random Walks in Biology, New Jersey: Princeton University Press.
Bomprezzi, R., P. Kovanen and R. Martin (2003). New approaches to investigating heterogeneity in complex traits. J. Med. Genet. 40, 553–559.
Carrero, G., E. Crawford, J. Th’ng, G. de Vries and M. J. Hendzel (2004). Quantification of protein-protein and protein-DNA interactions in vivo using fluorescence recovery after photobleaching. Methods Enzymol. 375, 415–442.
Carrero, G., D. McDonald, E. Crawford, G. de Vries and M. J. Hendzel (2003). Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods 29, 14–28.
Crank, J. (1975). The Mathematics of Diffusion, Oxford University Press.
Doi, M. and S. F. Edwards (1986). The Theory of Polymer Dynamics, New York: Oxford University Press.
Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L. Rothblum, R. Phair and T. Misteli (2002). A kinetic framework for a mammalian RNA polymerase in vivo. Science 298, 1623–1626.
Dundr, M. and T. Misteli (2001). Functional architecture in the cell nucleus. Biochem. J. 356, 297–310.
Eils, R. and C. Athale (2003). Computational imaging in cell biology. J. Cell Biol. 161, 477–481.
Hardt, S. L. (1978a). Aspects of diffusional transport in microorganisms, in Energetics and Structure of Halophilic Microorganisms, S. Caplan and M. Ginzburg (Eds), Amsterdam: Elsevier, pp. 591–597.
Hardt, S. L. (1978b). The diffusion transit time: a simple derivation. Bull. Math. Biol. 150, 591–597.
Hendzel, M. J., M. J. Kruhlak, N. A. MacLean, F. Boisvert, M. A. Lever and D. P. Bazett-Jones (2001). Compartmentalization of regulatory proteins in the cell nucleus. J. Steroid Biochem. Mol. Biol. 76, 9–21.
Hendzel, M., M. Lever, E. Crawford and J. Th’ng (2004). The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. (in press). (doi:10.1074/jbc.M400070200).
Howard, J. (2001). Mechanics of Motor Proteins and the Cytoskeleton, Massachusetts: Sinauer Associates. Inc.
Kruhlak, M. J., M. A. Lever, W. Fischle, E. Verdin, D. P. Bazzett-Jones and M. J. Hendzel (2000). Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady-state speckle compartments. J. Cell Biol. 150, 41–52.
Lever, M. A., J. P. Th’ng, X. Sung and M. J. Hendzel (2000). Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408, 873–876.
McDonald, D., G. Carrero, E. Crawford, C. Andrin, G. de Vries and M. J. Hendzel (2004). Nuclear beta actin exists in a dynamic equilibrium between filaments and monomers in mammalian cells (in preparation).
McGrath, J., Y. Tardy, C. Dewey, J. Meister and J. Hartwig (1998). Simultaneous measurements of actin filament turnover, filament fraction, and monomer diffusion in endothelial cells. Biophys. J. 75, 2070–2078.
Misteli, T. (2001). Protein dynamics: implications for nuclear architecture and gene expression. Science 291, 843–847.
Misteli, T., A. Gunjan, R. Hock, M. Bustin and D. T. Brown (2000). Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877–881.
Myers, R. H. (1986). Classical and Modern Regression with Applications, Boston: Duxbury Press.
Parada, L., J. Roix and T. Misteli (2003). An uncertainty principle in chromosome positioning. Trends Cell Biol. 13, 393–396.
Phair, R. D. and T. Misteli (2000). High mobility of proteins in the cell nucleus. Nature 404, 604–609.
Phair, R. D. and T. Misteli (2001). Kinetic modelling approaches to in vivo imaging. Nat. Rev. Mol. Cell Biol. 2, 898–907.
Politz, J. and A. Pombo (2002). Genomics meets nanoscience: probing genes and the cell nucleus at 10−9 meters. Genome Biol. 3, REPORTS4007.
Ronen, M., R. Rosenberg, B. Shraiman and U. Alon (2002). Assigning numbers to the arrows: parameterizing a gene regulation network by using accurate expression kinetics. Proc. Natl. Acad. Sci. USA 99, 10555–10560.
Stenoien, D. L., K. Patel, M. G. Mancini, M. Dutertre, C. L. Smith, B. W. O’Malley and M. A. Mancini (2001). FRAP reveals that mobility of oestrogen receptor-α is ligand-and proteasome-dependent. Nat. Cell. Biol. 3, 15–23.
Tardy, Y., J. McGrath, J. Hartwig and C. Dewey (1995). Interpreting photoactivated fluorescence microscopy measurements of steady-state actin dynamics. Biophys. J. 69, 1674–1682.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Carrero, G., Crawford, E., Hendzel, M.J. et al. Characterizing fluorescence recovery curves for nuclear proteins undergoing binding events. Bull. Math. Biol. 66, 1515–1545 (2004). https://doi.org/10.1016/j.bulm.2004.02.005
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1016/j.bulm.2004.02.005