Abstract
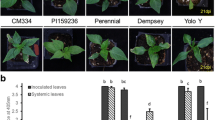
A resistant source (S-343) having monogenic dominant resistance to chilli leaf curl virus disease (ChiLCVD) has been identified at Punjab Agricultural University (PAU), Ludhiana. The F2 mapping population of 204 plants was derived from the cross MS-341 (susceptible) × S-343 (resistant) to identify the linked marker with the disease-resistant gene. Out of the 685 single-sequence repeats (SSRs) used, only 160 primers showed parental polymorphism. These 160 polymorphic primers were used for bulk segregant analysis and only eight SSR primers were able to differentiate the resistant and susceptible bulks. The linkage analysis revealed that the two markers CA 516044 and PAU-LC-343-1 were found linked with the disease-resistant gene covering a total distance of 15.7 centimorgan (cM). The two primers CA 516044 and PAU-LC-343-1 were found located on chromosome 6 of the pepper genome at a genetic distance of 6.8 cM and 8.9 cM, respectively, from the resistant gene. The validation of linked markers was performed using 26 resistant and susceptible genotypes developed at PAU, Ludhiana by former researchers. The validation of the primers revealed that there was a correlation between phenotypic and genotypic data of the used genotypes, and these markers can be used for the marker-assisted breeding procedures for transferring ChiLCVD resistance until the gene-based markers will be developed. The markers described in this study are the first-ever molecular markers identified as linked to the ChiLCVD-resistant gene.









Similar content being viewed by others
Change history
04 September 2020
The original article can be found online.
References
Ahmad A, Sharma A, Zehra SB, Kang SS, Bhat M, Hussain A (2016) Evaluation of chilli genotypes against Chilli leaf curl virus. Indian J Ecol 43:144–147
Aktar W, Sengupta D, Chowdhury A (2009) Impact of pesticides use in agriculture: their benefits and hazards. InterdiscipToxicol 2:1–2
Arjun K, Dhaliwal MS, Jindal SK, Fakrudin B (2018) Mapping of fruit length related QTLs in interspecific cross (Capsicum annuum L.×Capsicum galapagoenseHunz.) of chilli. Breed Sci 68:219–226
Aulakh PS, Dhaliwal MS, Jindal SK, Schafleitner R, Singh K (2016) Mapping of male sterility gene ms10 in chilli pepper (Capsicum annuum L.). Plant Breed 135:531–535
Barchi L, Bonnet J, Boudet C, Signoret P, Nagy I, Lanteri S, Palloix A, Lefebvre V (2007) A high-resolution, intraspecific linkage map of pepper (Capsicum annuum L.) and selection of reduced recombinant inbred line subsets for fast mapping. Genome 50:51–60
Ben-Chaim A, Borovsky Y, Falise M, Mazourek M, Kang BC, Paran L, Jahn M (2006) QTL analysis for capsaicinoid content in capsicum. TheorAppl Genet 113:1481–1490
Dhaliwal MS, Jindal SK, Cheema DS (2013) Punjab Sindhuri and Punjab Tej: new varieties of chilli. J Res Punjab AgricUniv 50:79–81
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. PhytoChem Bull 19:11–15
Ince AG, Karaca M, Onus AN (2010) Polymorphic microsatellite markers transferable across Capsicum species. Plant MolBiol Rep 28:285–291
Kang BC, Nahm SH, Huh JH, Yoo HS, Yu JW, Lee MH, Kim BD (2001) An interspecific (Capsicum annuum × C. chinese) F2 linkage map in pepper using RFLP and AFLP markers. Theor Appl Genet 102:531–539
Kenyon L, Kumar S, Tsai WS, Hughes JDA (2014) Virus diseases of peppers (Capsicum spp.) and their control. Advances in virus research, vol 904. Academic, New York, pp 297–335
Kim S, Park M, Yeom SI, Kim YM et al (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46:270–279
Kumar S, Kumar R, Kumar S, Singh AK, Singh M, Rai AB, Rai M (2011) Incidence of leaf curl disease on capsicum germplasm under field conditions. Indian J Agric Sci 8:187–189
Kumar RV, Singh AK, Singh AK, Yadav T, Basu S, Kushwaha N, Chattopadhyay B, Chakraborty S (2015) Complexity of begomovirus and betasatellite populations associated with chilli leaf curl disease in India. J Gen Virol 96:3143–3158
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Lee N (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lee JM, Nahm SH, Kim YM, Kim BD (2004) Characterization and molecular genetic mapping of microsatellite loci in pepper. TheorAppl Genet 108:619–627
Li W, Cheng J, Wu Z, Qin C, Tan S, Tang X, Cui J, Zhang L, Hu K (2015) An InDel-based linkage map of hot pepper (Capsicumannuum L.). Mol Breed 35:32
Livingstone KD, Lackney VK, Blauth JR, Wijk R, Jahn MK (1999) Genome mapping in capsicum and the evolution of genome structure in the Solanaceae. Genetics 152:1183–1202
Madden T (2013) The BLAST sequence analysis tool. In: The NCBI handbook [Internet], 2nd edn. https://www.ncbi.nlm.nih.gov/books/NBK153387/
Michelmore RW, Paran I, Kessel RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci 88:9828–9832
Minamiyama Y, Tsuro M, Hirai M (2006) An SSR-based linkage map of Capsicum annuum. Mol Breed 18:157–169
Pino J, Gonzalez M, Ceballos L, Centurion-Yah AR, Trujillo-Aguirred J, Latournerie- Moreno L, Sauri-Duch E (2007) Characterization of total capsaicinoidscolour and volatile compounds of Habanero chili pepper (Capsicum ChinenseJack.) cultivars grown in Yucatan. Food Chem 104:1682–1686
Prince JP, Pochard E, Tanskley SD (1993) Construction of a linkage map of pepper and a comparison of syntony with tomato. Genome 36:404–417
Senanayake DMJB, Mandal B, Lodha S (2007) Varma A (2007) First report of Chilli leaf curl virus affecting chilli in India. Plant Pathol 56:343
Senanayake DMJB, Varma A, Mandal BJ (2012) Virus–vector relationships, host range, detection and sequence comparison of chilli leaf curl virus associated with an epidemic of leaf curl disease of chilli in Jodhpur. Indian Phytopathol 160:146–155
Sharma A, Jindal SK, Thakur H (2018) Phenotypic classes of leaf curl virus disease severity for nursery screening in chilli pepper. Pl Dis Res 33:99–103
Sharma VK, Basu S, Chakraborty S (2015) RNAi mediated broad-spectrum transgenic resistance in Nicotiana benthamiana to chilli-infecting begomoviruses. Plant cell Rep 34:1389–1399
Tanskley SD, Bernatzky R, Lapitan NL, Prince JP (1988) Conservation of gene repertoire but not gene order in pepper and tomato. Proc Natl Acad Sci USA 85:6419–6423
Thakur H, Jindal SK, Sharma A, Dhaliwal MS (2017) Identification of resistant sources against chilli leaf curl virus disease. In: Indo-US symposium; Curbing whitefly—plant virus pandemics—The departure from pesticides to genomics solutions, Punjab Agricultural University, Ludhiana, Punjab, Abstr. No.: B14, p 72
Thakur H, Jindal SK, Sharma A, Dhaliwal MS (2018) Chilli leaf curl virus disease: a serious threat for chilli cultivation. J Plant Dis Protect 125:239–249
Thakur H, Jindal SK, Sharma A, Dhaliwal MS (2019) A monogenic dominant resistance for leaf curl virus disease in chilli pepper (Capsicum annuum L.). Crop Prot 116:115–120
Wang D, Bosland PW (2006) Thegenes of Capsicum. HortScience 41:1169–1187
Yi G, Lee JM, Lee S, Choi D, Kim BD (2006) Exploitation of pepper EST-SSRs and an SSR-based linkage map. Theor Appl Genet 114:113–130
Acknowledgements
The authors are thankful to Confederation of Indian Industry, Department of Science and Technology and Verdenta Hybrid Seeds Pvt. Ltd. for providing Prime Minister’s Fellowship to Hament Thakur.
Author information
Authors and Affiliations
Contributions
HT, SKJ, and AS conceived the experiments and wrote the original manuscript; SKJ, AS, and MSD revised the manuscript and supervise the experiments.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thakur, H., Jindal, S.K., Sharma, A. et al. Molecular mapping of dominant gene responsible for leaf curl virus resistance in chilli pepper (Capsicum annuum L.). 3 Biotech 10, 182 (2020). https://doi.org/10.1007/s13205-020-02168-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02168-7