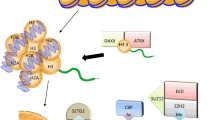
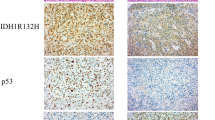
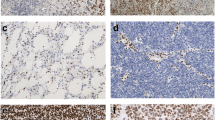
Abstract
Purpose of Review
This review will discuss the role of several key players in glioma classification and biology, namely isocitrate dehydrogenase 1 and 2 (IDH1/2), alpha thalassemia/mental retardation syndrome X-linked (ATRX), B-Raf (BRAF), telomerase reverse transcriptase (TERT), and H3K27M.
Recent Findings
IDH1/2 mutation delineates oligoden-droglioma, astrocytoma, and secondary glioblastoma (GBM) from primary GBM and lower-grade gliomas with biology similar to GBM. Additional mutations including TERT, 1p/19q, and ATRX further guide glioma classification and diagnosis, as well as pointing directions toward individualized treatments for these distinct molecular subtypes. ATRX and TERT mutations suggest the importance of telomere maintenance in gliomagenesis. BRAF alterations are key in certain low-grade gliomas and pediatric gliomas but rarely in high-grade gliomas in adults. Histone mutations (e.g., H3K27M) and their effect on chromatin modulation are novel mechanisms of cancer generation and uniquely seen in midline gliomas in children and young adults.
Summary
Over the past decade, a remarkable accumulation of knowledge from the genomic study of gliomas has led to reclassification of tumors, new understanding of oncogenic mechanisms, and novel treatment strategies.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance ••Of major importance
• Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–20. doi:10.1007/s00401-016-1545-1. This summary reviews the major changes to the 2016 WHO classification of tumors of the central nervous system and places new emphasis on the use of molecular information to classify gliomas.
Coons SW, Johnson PC, Scheithauer BW, Yates AJ, Pearl DK. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer. 1997;79(7):1381–93.
Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19(4):764–72. doi:10.1158/1078-0432.CCR-12-3002.
• Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–98. doi:10.1056/NEJMoa1402121. This study performed multi-platform sequencing of 293 LGG delineating mutational subtypes based on IDH, 1p/19q, and p53 status.
• Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–63. doi:10.1016/j.cell.2015.12.028. This study evaluated 1,122 diffuse gliomas showing the importance of ATRX and TERT in telomere maintenance of gliomas, as well as subtypes of IDH-mutant gliomas with poor prognosis versus subtypes of IDH-wild-type gliomas with good prognosis.
•• Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–508. doi:10.1056/NEJMoa1407279. This study performed multiplatform sequencing of 1087 gliomas delineating IDH mutation as an early mutational event and further classifying triple positive (IDH, 1p/19q, TERT mutant), ATRX/p53, TERT/p53, TERT-only, and triple negative (IDH, 1p/19q, TERT wild-type) tumors.
Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–12. doi:10.1126/science.1164382.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi:10.1016/j.ccr.2009.12.020.
Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–93. doi:10.1126/science.1239947.
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–73. doi:10.1056/NEJMoa0808710.
Karsy M, Sivakumar W, Neil JA, Moy F, Jensen RL. Meta-analysis of the effect of isocitrate dehydrogenase 1 and 2 mutation on glioblastoma prognosis. Contemp Neurosurg. 2015;37(5):1–5. doi:10.1097/01.cne.0000466893.11952.26.
Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi:10.1007/s00401-008-0455-2.
Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30(1):7–11. doi:10.1002/humu.20937.
Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–74. doi:10.1007/s00401-009-0561-9.
Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125(2):353–5. doi:10.1002/ijc.24379.
Kim W, Liau LM. IDH mutations in human glioma. Neurosurg Clin N Am. 2012;23(3):471–80. doi:10.1016/j.nec.2012.04.009.
Turkalp Z, Karamchandani J, Das S. IDH mutation in glioma: new insights and promises for the future. JAMA Neurol. 2014;71(10):1319–25. doi:10.1001/jamaneurol.2014.1205.
Bleeker FE, Atai NA, Lamba S, Jonker A, Rijkeboer D, Bosch KS, et al. The prognostic IDH1( R132) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol. 2010;119(4):487–94. doi:10.1007/s00401-010-0645-6.
Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–53. doi:10.2353/ajpath.2009.080958.
Mellai M, Piazzi A, Caldera V, Monzeglio O, Cassoni P, Valente G, et al. IDH1 and IDH2 mutations, immunohistochemistry and associations in a series of brain tumors. J Neurooncol. 2011;105(2):345–57. doi:10.1007/s11060-011-0596-3.
Duncan CG, Barwick BG, Jin G, Rago C, Kapoor-Vazirani P, Powell DR, et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res. 2012;22(12):2339–55. doi:10.1101/gr.132738.111.
Kranendijk M, Struys EA, van Schaftingen E, Gibson KM, Kanhai WA, van der Knaap MS, et al. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Science. 2010;330(6002):336. doi:10.1126/science.1192632.
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–44. doi:10.1038/nature08617.
Pietrak B, Zhao H, Qi H, Quinn C, Gao E, Boyer JG, et al. A tale of two subunits: how the neomorphic R132H IDH1 mutation enhances production of alphaHG. Biochemistry. 2011;50(21):4804–12. doi:10.1021/bi200499m.
Lee SM, Koh HJ, Park DC, Song BJ, Huh TL, Park JW. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002;32(11):1185–96.
Yan H, Bigner DD, Velculescu V, Parsons DW. Mutant metabolic enzymes are at the origin of gliomas. Cancer Res. 2009;69(24):9157–9. doi:10.1158/0008-5472.CAN-09-2650.
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–67. doi:10.1016/j.ccr.2010.11.015.
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi:10.1016/j.ccr.2010.12.014.
Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–8. doi:10.1038/nature10860.
Christensen BC, Smith AA, Zheng S, Koestler DC, Houseman EA, Marsit CJ, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst. 2011;103(2):143–53. doi:10.1093/jnci/djq497.
Laffaire J, Everhard S, Idbaih A, Criniere E, Marie Y, de Reynies A, et al. Methylation profiling identifies 2 groups of gliomas according to their tumorigenesis. Neuro Oncol. 2011;13(1):84–98. doi:10.1093/neuonc/noq110.
Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–22. doi:10.1016/j.ccr.2010.03.017.
Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–83. doi:10.1038/nature10866.
Fu Y, Zheng S, Zheng Y, Huang R, An N, Liang A, et al. Glioma derived isocitrate dehydrogenase-2 mutations induced up-regulation of HIF-1alpha and beta-catenin signaling: possible impact on glioma cell metastasis and chemo-resistance. Int J Biochem Cell Biol. 2012;44(5):770–5. doi:10.1016/j.biocel.2012.01.017.
Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483(7390):484–8. doi:10.1038/nature10898.
Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324(5924):261–5. doi:10.1126/science.1170944.
• Cohen A, Sato M, Aldape K, Mason CC, Alfaro-Munoz K, Heathcock L, et al. DNA copy number analysis of Grade II-III and Grade IV gliomas reveals differences in molecular ontogeny including chromothripsis associated with IDH mutation status. Acta Neuropathol Commun. 2015;3:34. doi:10.1186/s40478-015-0213-3. This study showed that IDH conferred improved survival for both LGG and HGG. In addition, IDH mutation was associated with increased chromosomal instability, namely chromothripsis.
• Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–4. doi:10.1038/nature16490. This study showed a novel role for IDH mutation where hypermethylation at cohesion and CCTC-binding factor (CTCF)-binding sites within the genome could cause disruption of a methylation-sensitive insulator protein and aberrant gene expression.
Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224(3):334–43. doi:10.1002/path.2913.
Pansuriya TC, van Eijk R, d’Adamo P, van Ruler MA, Kuijjer ML, Oosting J, et al. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet. 2011;43(12):1256–61. doi:10.1038/ng.1004.
Karsy M, Neil JA, Guan J, Mahan MA, Colman H, Jensen RL. A practical review of prognostic correlations of molecular biomarkers in glioblastoma. Neurosurg Focus. 2015;38(3):E4. doi:10.3171/2015.1.FOCUS14755.
Labussiere M, Idbaih A, Wang XW, Marie Y, Boisselier B, Falet C, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74(23):1886–90. doi:10.1212/WNL.0b013e3181e1cf3a.
Wakimoto H, Tanaka S, Curry WT, Loebel F, Zhao D, Tateishi K, et al. Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin Cancer Res. 2014;20(11):2898–909. doi:10.1158/1078-0432.CCR-13-3052.
Chen R, Ravindra VM, Cohen AL, Jensen RL, Salzman KL, Prescot AP, et al. Molecular features assisting in diagnosis, surgery, and treatment decision making in low-grade gliomas. Neurosurg Focus. 2015;38(3):E2. doi:10.3171/2015.1.FOCUS14745.
Tabouret E, Nguyen AT, Dehais C, Carpentier C, Ducray F, Idbaih A, et al. Prognostic impact of the 2016 WHO classification of diffuse gliomas in the French POLA cohort. Acta Neuropathol. 2016;132(4):625–34. doi:10.1007/s00401-016-1611-8.
Sun H, Yin L, Li S, Han S, Song G, Liu N, et al. Prognostic significance of IDH mutation in adult low-grade gliomas: a meta-analysis. J Neurooncol. 2013;113(2):277–84. doi:10.1007/s11060-013-1107-5.
Cairncross JG, Wang M, Jenkins RB, Shaw EG, Giannini C, Brachman DG, et al. Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol. 2014;32(8):783–90. doi:10.1200/JCO.2013.49.3726.
van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344–50. doi:10.1200/JCO.2012.43.2229.
Elkhaled A, Jalbert LE, Phillips JJ, Yoshihara HA, Parvataneni R, Srinivasan R, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4(116):116ra5. doi:10.1126/scitranslmed.3002796.
Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. 2-Hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18(4):624–9. doi:10.1038/nm.2682.
Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107(1):197–205. doi:10.1007/s11060-011-0737-8.
Andronesi OC, Loebel F, Bogner W, Marjanska M, Vander Heiden MG, Iafrate AJ, et al. Treatment response assessment in IDH-mutant glioma patients by noninvasive 3D functional spectroscopic mapping of 2-hydroxyglutarate. Clin Cancer Res. 2016;22(7):1632–41. doi:10.1158/1078-0432.CCR-15-0656.
Beiko J, Suki D, Hess KR, Fox BD, Cheung V, Cabral M, et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol. 2014;16(1):81–91. doi:10.1093/neuonc/not159.
Fujii T, Khawaja MR, DiNardo CD, Atkins JT, Janku F. Targeting isocitrate dehydrogenase (IDH) in cancer. Discov Med. 2016;21(117):373–80.
Popovici-Muller J, Saunders JO, Salituro FG, Travins JM, Yan S, Zhao F, et al. Discovery of the first potent inhibitors of mutant IDH1 that lower tumor 2-HG in vivo. ACS Med Chem Lett. 2012;3(10):850–5. doi:10.1021/ml300225h.
Chaturvedi A, Araujo Cruz MM, Jyotsana N, Sharma A, Yun H, Gorlich K, et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122(16):2877–87. doi:10.1182/blood-2013-03-491571.
Li L, Paz AC, Wilky BA, Johnson B, Galoian K, Rosenberg A, et al. Treatment with a small molecule mutant IDH1 inhibitor suppresses tumorigenic activity and decreases production of the oncometabolite 2-hydroxyglutarate in human chondrosarcoma cells. PLoS One. 2015;10(9):e0133813. doi:10.1371/journal.pone.0133813.
Burris H, Mellinghoff I, Maher E, Wen P, Beeram M, Touat M, et al. Abstract PL04-05: the first reported results of AG-120, a first-in-class, potent inhibitor of the IDH1 mutant protein, in a phase I study of patients with advanced IDH1-mutant solid tumors, including gliomas. Mol Cancer Ther. 2015;14(12 Supplement 2):PL04-5-PL-5. doi:10.1158/1535-7163.targ-15-pl04-05.
DeBotton S, Pollyea D, Stein E, Dinardo C, Fathi A, Roboz G, et al. Clinical safety and activity of AG-120, a first-in-class, potent inhibitor of the IDH1 mutant protein, in a phase 1 study of patients with advanced IDH1-mutant hematologic malignancies. 20th Annual Meeting of European Hematology Association; 2015. Vienna, Austria.
Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–6. doi:10.1126/science.1234769.
Yen K, Wang F, Travins J, Chen Y, Yang H, Straley K, et al. AG-221 offers a survival advantage in a primary human IDH2 mutant AML xenograft model. Blood. 2013;122(21):240.
Dinardo C, Stein E, Altman J, Collins R, Deangelo D, Fathi A, et al. AG-221, an oral, selective, first-in-class, potent inhibitor of the IDH2 mutant enzyme, induced durable responses in a phase 1 study of IDH2 mutation-positive advanced hematologic malignancies. 20th Annual Meeting of European Hematology Association; 2015. Vienna.
Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–30. doi:10.1126/science.1236062.
Pellegatta S, Valletta L, Corbetta C, Patane M, Zucca I, Riccardi Sirtori F, et al. Effective immuno-targeting of the IDH1 mutation R132H in a murine model of intracranial glioma. Acta Neuropathol Commun. 2015;3:4. doi:10.1186/s40478-014-0180-0.
Schumacher T, Bunse L, Pusch S, Sahm F, Wiestler B, Quandt J, et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–7. doi:10.1038/nature13387.
Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(6041):425. doi:10.1126/science.1207313.
Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–31. doi:10.1038/nature10833.
• Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–50. doi:10.1038/ng.2938. Comprehensive genomic evaluation of 127 pediatric gliomas showing alterations in histone H3, p53, and ATRX in both DIPGs and other HGG. Mutations in PI3K signaling, histone modification, and cell cycle genes were common.
Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–22. doi:10.18632/oncotarget.588.
Kannan K, Inagaki A, Silber J, Gorovets D, Zhang J, Kastenhuber ER, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3(10):1194–203. doi:10.18632/oncotarget.689.
Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124(5):615–25. doi:10.1007/s00401-012-1031-3.
Ikemura M, Shibahara J, Mukasa A, Takayanagi S, Aihara K, Saito N, et al. Utility of ATRX immunohistochemistry in diagnosis of adult diffuse gliomas. Histopathology. 2016;69(2):260–7. doi:10.1111/his.12927.
Venneti S, Huse JT. The evolving molecular genetics of low-grade glioma. Adv Anat Pathol. 2015;22(2):94–101. doi:10.1097/PAP.0000000000000049.
Cai J, Zhu P, Zhang C, Li Q, Wang Z, Li G, et al. Detection of ATRX and IDH1-R132H immunohistochemistry in the progression of 211 paired gliomas. Oncotarget. 2016;7(13):16384–95. doi:10.18632/oncotarget.7650.
Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107(32):14075–80. doi:10.1073/pnas.1008850107.
Abedalthagafi M, Phillips JJ, Kim GE, Mueller S, Haas-Kogen DA, Marshall RE, et al. The alternative lengthening of telomere phenotype is significantly associated with loss of ATRX expression in high-grade pediatric and adult astrocytomas: a multi-institutional study of 214 astrocytomas. Mod Pathol. 2013;26(11):1425–32. doi:10.1038/modpathol.2013.90.
Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179(4):1608–15. doi:10.1016/j.ajpath.2011.06.018.
Eid R, Demattei MV, Episkopou H, Auge-Gouillou C, Decottignies A, Grandin N, et al. Genetic inactivation of ATRX leads to a decrease in the amount of telomeric cohesin and level of telomere transcription in human glioma cells. Mol Cell Biol. 2015;35(16):2818–30. doi:10.1128/MCB.01317-14.
Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347(6219):273–7. doi:10.1126/science.1257216.
Cai J, Chen J, Zhang W, Yang P, Zhang C, Li M, et al. Loss of ATRX, associated with DNA methylation pattern of chromosome end, impacted biological behaviors of astrocytic tumors. Oncotarget. 2015;6(20):18105–15. doi:10.18632/oncotarget.3906.
Wiestler B, Capper D, Holland-Letz T, Korshunov A, von Deimling A, Pfister SM, et al. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126(3):443–51. doi:10.1007/s00401-013-1156-z.
Reuss DE, Sahm F, Schrimpf D, Wiestler B, Capper D, Koelsche C, et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an "integrated" diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol. 2015;129(1):133–46. doi:10.1007/s00401-014-1370-3.
Chi AS, Batchelor TT, Yang D, Dias-Santagata D, Borger DR, Ellisen LW, et al. BRAF V600E mutation identifies a subset of low-grade diffusely infiltrating gliomas in adults. J Clin Oncol. 2013;31(14):e233–6. doi:10.1200/JCO.2012.46.0220.
Dahiya S, Emnett RJ, Haydon DH, Leonard JR, Phillips JJ, Perry A, et al. BRAF-V600E mutation in pediatric and adult glioblastoma. Neuro Oncol. 2014;16(2):318–9. doi:10.1093/neuonc/not146.
Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. doi:10.1007/s00401-011-0802-6.
Lee D, Cho YH, Kang SY, Yoon N, Sung CO, Suh YL. BRAF V600E mutations are frequent in dysembryoplastic neuroepithelial tumors and subependymal giant cell astrocytomas. J Surg Oncol. 2015;111(3):359–64. doi:10.1002/jso.23822.
Bar EE, Lin A, Tihan T, Burger PC, Eberhart CG. Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. J Neuropathol Exp Neurol. 2008;67(9):878–87. doi:10.1097/NEN.0b013e3181845622.
Jacob K, Albrecht S, Sollier C, Faury D, Sader E, Montpetit A, et al. Duplication of 7q34 is specific to juvenile pilocytic astrocytomas and a hallmark of cerebellar and optic pathway tumours. Br J Cancer. 2009;101(4):722–33. doi:10.1038/sj.bjc.6605179.
Pfister S, Janzarik WG, Remke M, Ernst A, Werft W, Becker N, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118(5):1739–49. doi:10.1172/JCI33656.
Kleinschmidt-DeMasters BK, Aisner DL, Birks DK, Foreman NK. Epithelioid GBMs show a high percentage of BRAF V600E mutation. Am J Surg Pathol. 2013;37(5):685–98. doi:10.1097/PAS.0b013e31827f9c5e.
• Mistry M, Zhukova N, Merico D, Rakopoulos P, Krishnatry R, Shago M, et al. BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high-grade glioma. J Clin Oncol. 2015;33(9):1015–22. doi:10.1200/JCO.2014.58.3922. This study evaluated 886 cases of pediatric LGG showing that BRAF-V600E mutations were common and important for progression to secondary HGG.
Robinson JP, VanBrocklin MW, Guilbeault AR, Signorelli DL, Brandner S, Holmen SL. Activated BRAF induces gliomas in mice when combined with Ink4a/Arf loss or Akt activation. Oncogene. 2010;29(3):335–44. doi:10.1038/onc.2009.333.
Shin CH, Grossmann AH, Holmen SL, Robinson JP. The BRAF kinase domain promotes the development of gliomas in vivo. Genes Cancer. 2015;6(1-2):9–18. doi:10.18632/genesandcancer.48.
Raabe EH, Lim KS, Kim JM, Meeker A, Mao XG, Nikkhah G, et al. BRAF activation induces transformation and then senescence in human neural stem cells: a pilocytic astrocytoma model. Clin Cancer Res. 2011;17(11):3590–9. doi:10.1158/1078-0432.CCR-10-3349.
Aguilera D, Janss A, Mazewski C, Castellino RC, Schniederjan M, Hayes L, et al. Successful retreatment of a child with a refractory brainstem ganglioglioma with vemurafenib. Pediatr Blood Cancer. 2016;63(3):541–3. doi:10.1002/pbc.25787.
Bautista F, Paci A, Minard-Colin V, Dufour C, Grill J, Lacroix L, et al. Vemurafenib in pediatric patients with BRAFV600E mutated high-grade gliomas. Pediatr Blood Cancer. 2014;61(6):1101–3. doi:10.1002/pbc.24891.
Chamberlain MC. Salvage therapy with BRAF inhibitors for recurrent pleomorphic xanthoastrocytoma: a retrospective case series. J Neurooncol. 2013;114(2):237–40. doi:10.1007/s11060-013-1176-5.
del Bufalo F, Carai A, Figa-Talamanca L, Pettorini B, Mallucci C, Giangaspero F, et al. Response of recurrent BRAFV600E mutated ganglioglioma to vemurafenib as single agent. J Transl Med. 2014;12:356. doi:10.1186/s12967-014-0356-1.
Lee EQ, Ruland S, LeBoeuf NR, Wen PY, Santagata S. Successful treatment of a progressive BRAF V600E-mutated anaplastic pleomorphic xanthoastrocytoma with vemurafenib monotherapy. J Clin Oncol. 2016;34(10):e87–9. doi:10.1200/JCO.2013.51.1766.
Robinson GW, Orr BA, Gajjar A. Complete clinical regression of a BRAF V600E-mutant pediatric glioblastoma multiforme after BRAF inhibitor therapy. BMC Cancer. 2014;14:258. doi:10.1186/1471-2407-14-258.
Rush S, Foreman N, Liu A. Brainstem ganglioglioma successfully treated with vemurafenib. J Clin Oncol. 2013;31(10):e159–60. doi:10.1200/JCO.2012.44.1568.
Usubalieva A, Pierson CR, Kavran CA, Huntoon K, Kryvenko ON, Mayer TG, et al. Primary meningeal pleomorphic xanthoastrocytoma with anaplastic features: a report of 2 cases, one with BRAF(V600E) mutation and clinical response to the BRAF inhibitor dabrafenib. J Neuropathol Exp Neurol. 2015;74(10):960–9. doi:10.1097/NEN.0000000000000240.
Lang SS. The role of BRAF-targeted therapy in astrocytomas: a review. Neurosurgery. 2013;60 Suppl 1:110–2. doi:10.1227/01.neu.0000430768.25844.4d.
Sievert AJ, Lang SS, Boucher KL, Madsen PJ, Slaunwhite E, Choudhari N, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A. 2013;110(15):5957–62. doi:10.1073/pnas.1219232110.
Arndt GM, MacKenzie KL. New prospects for targeting telomerase beyond the telomere. Nat Rev Cancer. 2016;16(8):508–24. doi:10.1038/nrc.2016.55.
Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz Jr LA, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6. doi:10.1073/pnas.1303607110.
Shervington A, Patel R, Lu C, Cruickshanks N, Lea R, Roberts G, et al. Telomerase subunits expression variation between biopsy samples and cell lines derived from malignant glioma. Brain Res. 2007;1134(1):45–52. doi:10.1016/j.brainres.2006.11.093.
• Walsh KM, Codd V, Smirnov IV, Rice T, Decker PA, Hansen HM, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46(7):731–5. doi:10.1038/ng.3004. An analysis of SNPs in gliomas showed a high likelihood for alterations near TERC and TERT, which were implicated in telomere maintenance.
Geng P, Zhao X, Ou J, Li J, Sa R, Liang H. TERT genetic mutations as prognostic marker in glioma. Mol Neurobiol. In press. doi:10.1007/s12035-016-9930-2.
Yuan Y, Qi C, Maling G, Xiang W, Yanhui L, Ruofei L, et al. TERT mutation in glioma: frequency, prognosis and risk. J Clin Neurosci. 2016;26:57–62. doi:10.1016/j.jocn.2015.05.066.
Labussiere M, Di Stefano AL, Gleize V, Boisselier B, Giry M, Mangesius S, et al. TERT promoter mutations in gliomas, genetic associations and clinico-pathological correlations. Br J Cancer. 2014;111(10):2024–32. doi:10.1038/bjc.2014.538.
Filbin MG, Suva ML. Gliomas genomics and epigenomics: arriving at the start and knowing it for the first time. Annu Rev Pathol. 2016;11:497–521. doi:10.1146/annurev-pathol-012615-044208.
Lulla RR, Saratsis AM, Hashizume R. Mutations in chromatin machinery and pediatric high-grade glioma. Sci Adv. 2016;2(3):e1501354. doi:10.1126/sciadv.1501354.
Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014;14(10). doi:10.1038/nrc3811.
Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–3. doi:10.1038/ng.1102.
Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–37. doi:10.1016/j.ccr.2012.08.024.
Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet. 2014;46(5):462–6. doi:10.1038/ng.2950.
Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–61. doi:10.1126/science.1232245.
Bax DA, Mackay A, Little SE, Carvalho D, Viana-Pereira M, Tamber N, et al. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res. 2010;16(13):3368–77. doi:10.1158/1078-0432.CCR-10-0438.
Taylor KR, Mackay A, Truffaux N, Butterfield YS, Morozova O, Philippe C, et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet. 2014;46(5):457–61. doi:10.1038/ng.2925.
Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21(6):555–9. doi:10.1038/nm.3855.
Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20(12):1394–6. doi:10.1038/nm.3716.
Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–8. doi:10.1038/nature11262.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–401. doi:10.1126/science.1254257.
Efroni I, Ip PL, Nawy T, Mello A, Birnbaum KD. Quantification of cell identity from single-cell gene expression profiles. Genome Biol. 2015;16:9. doi:10.1186/s13059-015-0580-x.
Meyer M, Reimand J, Lan X, Head R, Zhu X, Kushida M, et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A. 2015;112(3):851–6. doi:10.1073/pnas.1320611111.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Michael Karsy and Jian Guan declare that they have no conflict of interest.
Adam L. Cohen reports grants from Novartis, Genentech, Merrimack, and BMS.
Randy L. Jensen reports personal fees from Pharmacokinesis and Medtronic.
Howard Colman has received advisory board fees from Roche, Genentech, Upsher-Smith, Oxigene, CytRx, Omniox, and Insys, and grants from Plexxikon and Newlink Genetics.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Neuro-Oncology
Rights and permissions
About this article
Cite this article
Karsy, M., Guan, J., Cohen, A.L. et al. New Molecular Considerations for Glioma: IDH, ATRX, BRAF, TERT, H3 K27M. Curr Neurol Neurosci Rep 17, 19 (2017). https://doi.org/10.1007/s11910-017-0722-5
Published:
DOI: https://doi.org/10.1007/s11910-017-0722-5