Abstract
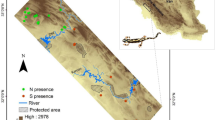
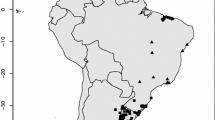
Environmental shifts are projected to lead to expansions, contractions and shifts in the geographic range of species in response to climate change. Most distribution forecasting methods assume that species respond to the environment as an undifferentiated entity along their entire distribution. However, environmental heterogeneity along a species’ range plays a key role in driving the evolutionary trajectories of populations and species by determining intraspecific variation in morphological, physiological and life history traits. Therefore, for widely distributed species with phylogeographic structure, the construction of SDM where lineages are pooled within the species level might not accurately describe the niche of sets of lineages that are adapted to different climatic conditions. Pleurodema thaul is an endemic frog distributed from the Atacama Desert to the temperate rainforest in southern Chile and Argentina. Along this latitudinal gradient, populations exhibit variation in thermal physiology, life history traits and phylogenetic structure. We evaluated whether evolutionary lineages differ in their environmental and geographic niches and determined how phylogenetic, environmental and geographical distances covary between clades. Our results indicate that: (i) all clades of this species differ in their estimated niches, (ii) there is little overlap between geographic areas and environmental niches estimated for each clade of P. thaul and (iii) phylogenetic distances among clades of P. thaul are not correlated with environmental nor geographical distances. The development of models restricted to species’ genetic lineages can become an important first step to determine how environmental changes are going to affect the distribution of species.





Similar content being viewed by others
Abbreviations
- SDM:
-
Species distribution model
- RF:
-
Random forest
- CV:
-
Cross-validation
- AUC:
-
Area under the curve
- ROC:
-
Receiver operating characteristic
- TSS:
-
True skill statistic
- PCA:
-
Principal components analysis
- MVE:
-
Minimum–volume ellipsoid
References
Allouche, O., Tsoar, A., & Kadmon, R. (2006). Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). Journal of Applied Ecology,43, 1223–1232.
Aurenhammer, F., & Klein, R. (2000). Voronoi diagrams. In J. R. Sack & J. Urrutia (Eds.), Handbook of computational geometry. Amsterdam: Elsevier.
Avise, J. C. (2000). Phylogeography: the history and formation of species. Massachusetts: Harvard University Press.
Barria, A. M., & Bacigalupe, L. D. (2017). Intraspecific geographic variation in thermal limits and acclimatory capacity in a wide distributed endemic frog. Journal of Thermal Biology,69, 254–260.
Beaumont, L. J., Pitman, A., Perkins, S., Zimmermann, N. E., Yoccoz, N. G., & Thuiller, W. (2011). Impacts of climate change on the world's most exceptional ecoregions. Proceeding of the National Academy of Sciences of the USA,108, 2306–2311.
Beebee, T. J. C. (1996). Ecology and conservation of amphibians. London: Chapman & Hall.
Bell, G. (2017). Evolutionary rescue. Annual Review of Ecology, Evolution and Systematics,48, 605–627.
Blonder, B., Lamanna, C., Violle, C., & Enquist, B. J. (2014). The n-dimensional hypervolume. Global Ecology and Biogeography,23, 595–609.
Bocedi, G., Atkins, K. E., Liao, J., Henry, R. C., Travis, J. M., & Hellmann, J. J. (2013). Effects of local adaptation and interspecific competition on species’ responses to climate change. Annals of the New York Academy of Sciences,1297, 83–97.
Booth, T. H., Nix, H. A., Busby, J. R., & Hutchinson, M. F. (2014). bioclim: The first species distribution modelling package, its early applications and relevance to most current MaxEnt studies. Diversity and Distributions,20, 1–9.
Bosch, S., Tyberghein, L., & De Clerck, O. (2017). sdmpredictors: An R package for species distribution modelling predictor datasets. Marine Species distributions: From data to predictive models, 49.
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: A software platform for Bayesian evolutionary analysis. PLOS Computational Biology,10(4), e1003537.
Bradshaw, A. D. (1991). The Croonian lecture. Genostasis and the limits to evolution. Philosophical Transactions of the Royal Society B: Biological Sciences,333(1267), 289–305.
Breiman, L. (2001). Random forests. Machine Learning,45, 5–32.
Charmantier, A., McCleery, R. H., Cole, L. R., Perrins, C., Kruuk, E. B., & Sheldon, B. C. (2008). Adaptive phenotypic plasticity in response to climate change in a wild bird population. Science,320(5877), 800–803.
Chen, I. C., Hill, J. K., Ohlemüller, R., Roy, D. B., & Thomas, C. D. (2011). Rapid range shifts of species associated with high levels of climate warming. Science,333(6045), 1024–1026.
Chessel, D., Dufour, A. B., & Thioulouse, J. (2004). The ade4 package-I-One-table methods. R News,4(1), 5–10.
Cheung, W. W. L., Lam, V. W., Sarmiento, J. L., Kearney, K., Watson, R. E. G., Zeller, D., et al. (2010). Large scale redistribution of maximum fisheries catch potential in the global ocean under climate change. Global Change Biology,16(1), 24–35.
Correa, C., Lobos, G., Pastenes, L., & Méndez, M. A. (2008). Invasive Pleurodema thaul from Robinson Crusoe Island: molecular identification of its geographic origin and comments on the phylogeographic structure of this species in mainland Chile. Journal of Herpetology,18(2), 77–82.
Cutler, D. R., Edwards, T. C., Beard, K. H., Cutler, A., Hess, K. T., Gibson, J., et al. (2007). Random forests for classification in ecology. Ecology,88, 2783–2792.
D’Amen, M., Zimmermann, N. E., & Pearman, P. B. (2013). Conservation of phylogeographic lineages under climate change. Global Ecology and Biogeography,22(1), 93–104.
Dormann, C. F., Elith, J., Bacher, S., Buchmann, C., Carl, G., Carre, G., et al. (2013). Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography,36, 27–46.
Dray, S., Dufour, A. B., & Chessel, D. (2007). The ade4 package-II: Two-table and K-table methods. R news,7(2), 47–52.
Dray, S., & Dufour, A. B. (2007). The ade4 package: Implementing the duality diagram for ecologists. Journal of Statistical Software,22(4), 1–20.
Dyer, R. J., Nason, J. D., & Garrick, R. C. (2010). Landscape modelling of gene flow: Improved power using conditional genetic distance derived from the topology of population networks. Molecular Ecology,19, 3746–3759.
Elith, J., Graham, C. H., Anderson, R. P., Dudík, M., Ferrier, S., Guisan, A., et al. (2006). Novel methods improve prediction of species distributions from occurrence data. Ecography,29, 129–151.
Elith, J., & Leathwick, J. R. (2009). Species distribution models: Ecological explanation and prediction across space and time. Annual Review of Ecology, Evolution and Systematics,40, 677–697.
Faivovich, J., Ferraro, D. P., Basso, N. G., Haddad, C. F., Rodrigues, M. T., Wheeler, W. C., et al. (2012). A phylogenetic analysis of Pleurodema (Anura: Leptodactylidae: Leiuperinae) based on mitochondrial and nuclear gene sequences, with comments on the evolution of anuran foam nests. Cladistics,28(5), 460–482.
Fangue, N. A., Hofmeister, M., & Schulte, P. M. (2006). Intraspecific variation in thermal tolerance and heat shock protein gene expression in common killifish Fundulus heteroclitus. Journal of Experimental Biology,209, 2859–2872.
Fick, S. E., & Hijmans, R. J. (2017). WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology,37, 4302–4315.
Fielding, A. H., & Bell, J. F. (1997). A review of methods for the assessment of prediction errors in conservation presence/absence models. Environmental Conservation,24(1), 38–49.
Franklin, J. (2010). Mapping species distributions: Spatial inference and prediction. Cambridge: University Press.
Freeland, J. R., Biss, P., Conrad, K. F., & Silvertown, J. (2010). Selection pressures have caused genome wide population differentiation of Anthoxanthum odoratum despite the potential for high gene flow. Journal of Evolutionary Biology,23(4), 776–782.
Freeman, E. A., & Moisen, G. (2008). PresenceAbsence: An R package for presence-absence model analysis. Journal of Statistical Software,23(11), 1–31.
Garant, D., Kruuk, L. E. B., Wilkin, T. A., McCleery, R. H., & Sheldon, B. C. (2005). Evolution driven by differential dispersal within a wild bird population. Nature,433, 60–65.
Godoy, B. S., Camargos, L. M., & Lodi, S. (2018). When phylogeny and ecology meet: Modeling the occurrence of Trichoptera with environmental and phylogenetic data. Ecology and Evolution,8(11), 5313–5322.
Gotelli, N. J., & Stanton-Geddes, J. (2015). Climate change, genetic markers and species distribution modelling. Journal of Biogeography,42(9), 1577–1585.
Guisan, A., & Thuiller, W. (2005). Predicting species distribution: Offering more than simple habitat models. Ecology Letters,8, 993–1009.
Guisan, A., Tingley, R., Baumgartner, J. B., Naujokaitis-Lewis, I., Sutcliffe, P. R., Tulloch, A. I., et al. (2013). Predicting species distributions for conservation decisions. Ecology Letters,16(12), 1424–1435.
Guisan, A., & Zimmermann, N. E. (2000). Predictive habitat distribution models in ecology. Ecological Modelling,135, 147–186.
Ikeda, D. H., Max, T. L., Allan, G. J., Lau, M. K., Shuster, S. M., & Whitham, T. G. (2017). Genetically informed ecological niche models improve climate change predictions. Global Change Biology,23(1), 164–176.
IPBES. (2019). Summary for policymakers of the global assessment report on biodiversity and ecosystem services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services. Bonn: IPBES secretariat.
Iturra-Cid, M. S., Ortiz, J. C., & Ibargüengoytía, N. R. (2010). Age, size, and growth of the chilean frog Pleurodema thaul (Anura: Leiuperidae): Latitudinal and altitudinal effects. Copeia,4, 609–617.
Jaccard, P. (1901). Distribution de la flore alpine dans le Bassin des Drouces et dans quelques regions voisines. Bulletin de la Société Vaudoise des Sciences Naturelles,37(140), 241–272.
Jackson, A. L., Parnell, A. C., Inger, R., & Bearhop, S. (2011). Comparing isotopic niche widths among and within communities: SIBER—Stable Isotope Bayesian Ellipses in R. Journal of Animal Ecology,80(3), 595–602.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., & Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods,14, 587–589.
Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution,30(4), 772–780.
Kearney, M., Porter, W. P., Williams, C., Ritchie, S., & Hoffmann, A. A. (2009). Integrating biophysical models and evolutionary theory to predict climatic impacts on species’ ranges: The dengue mosquito Aedes aegipti in Australia. Functional Ecology,23, 528–538.
Kuhn, M. (2008). Building predictive models in R using the caret package. Journal of Statistical Software,28, 1–26.
Kuhn, M., & Johnson, K. (2013). Applied predictive modeling. New York: Springer.
Lee-Yaw, J. A., & Irwin, D. E. (2015). The importance (or lack thereof) of niche divergence to the maintenance of a northern species complex: the case of the long toed salamander (Ambystoma macrodactylum, Baird). Journal of Evolutionary Biology,28(4), 917–930.
Lenoir, J., Gégout, J. C., Marquet, P. A., de Ruffray, P., & Brisse, H. (2008). A significant upward shift in plant species optimum elevation during the 20th century. Science,320(5884), 1768–1771.
Luebert, F., & Pliscoff, P. (2006). Sinopsis bioclimática y vegetacional de Chile. Chile: Editorial Universitaria.
Manel, S., Dias, J. M., & Ormerod, S. J. (1999). Comparing discriminant analysis, neural networks and logistic regression for predicting species distributions: A case study with a Himalayan river bird. Ecological Modelling,120, 337–347.
Merilä, J., & Hendry, A. P. (2014). Climate change, adaptation, and phenotypic plasticity: the problem and the evidence. Evolutionary Applications,7(1), 1–14.
Millennium Ecosystem Assessment Program. (2005). Ecosystems and human well-being. Washington, DC: Island Press.
Morales-Castilla, I., Davies, T. J., Pearse, W. D., & Peres-Neto, P. (2017). Combining phylogeny and co-occurrence to improve single species distribution models. Global Ecology and Biogeography,26(6), 740–752.
Moran, E. V., & Ormond, R. A. (2015). Simulating the interacting effects of intraspecific variation, disturbance, and competition on climate-driven range shifts in trees. PLoS ONE,10(11), e0142369.
Moyes, K., Nussey, D. H., Clements, M. N., Guinness, F. E., Morris, A., Morris, S., et al. (2011). Advancing breeding phenology in response to environmental change in a wild red deer population. Global Change Biology,17(7), 2455–2469.
Norris, J. R., Jackson, S. T., & Betancourt, J. L. (2006). Classification tree and minimum-volume ellipsoid analyses of the distribution of ponderosa pine in the western USA. Journal of Biogeography,33(2), 342–360.
Okabe, A. (2016). Spatial tessellations. International Encyclopedia of Geography: People, the Earth, Environment and Technology, pp. 1–11.
Osborne, P. E., Suátez-Seoane, S., & Alonso, J. C. (2007). Behavioural mechanisms that undermine species envelope models: The causes of patchiness in the distribution of great bustards Otis tarda in Spain. Ecography,30, 819–828.
Paradis, E., & Schliep, K. (2018). ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics,35(3), 526–528.
Pearman, P. B., Guisan, A., Broennimann, O., & Randin, C. F. (2008). Niche dynamics in space and time. Trends in Ecology and Evolution,23(3), 149–158.
Pearman, P. B., D'Amen, M., Graham, C. H., Thuiller, W., & Zimmermann, N. E. (2010). Within-taxon niche structure: Niche conservatism, divergence and predicted effects of climate change. Ecography,33(6), 990–1003.
Pereira, H. M., Leadley, P. W., Proença, V., Alkemade, R., Scharlemann, J. P. W., Fernandez-Manjarrés, J. F., et al. (2010). Scenarios for global biodiversity in the 21st century. Science,330(6010), 1496–1501.
Peterson, A. T., & Soberón, J. (2012). Species distribution modeling and ecological niche modeling: Getting the concepts right. Natureza & Conservação,10(2), 102–107.
Peterson, A. T., Soberón, J., & Sánchez-Cordero, V. (1999). Conservatism of ecological niches in evolutionary time. Science,285(5431), 1265–1267.
Peterson, A. T., Soberón, J., Pearson, R. G., Anderson, R. P., Martínez-Meyer, E., Nakamura, M., et al. (2011). Ecological niches and geographic distributions. monographs in population biology. Princeton: Princeton University Press.
Portner, H. O., & Knust, R. (2007). Climate change affects marine fishes through the oxygen limitation of thermal tolerance. Science,315(5808), 95–97.
Postma, E., & van Noordwijk, A. J. (2005). Gene flow maintains a large genetic difference in clutch size at a small spatial scale. Nature,433(7021), 65.
QGIS Development Team. (2020). QGIS geographic information system. Open source geospatial foundation project. https://qgis.osgeo.org.
R Core Team (2017) R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. https://www.R-project.org/.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., & Suchard, M. A. (2018). Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology,67, 901.
Randin, C. F., Engler, R., Normand, S., Zappa, M., Zimmermann, N. E., Pearman, P. B., et al. (2009). Climate change and plant distribution: local models predict high elevation persistence. Global Change Biology,15(6), 1557–1569.
Sarricolea, P., Herrera-Ossandon, M., & Meseguer-Ruiz, Ó. (2017). Climatic regionalisation of continental Chile. Journal of Maps,13(2), 66–73.
Schoville, S. D., Bonin, A., François, O., Lobreaux, S., Melodelima, C., & Manel, S. (2012). Adaptive genetic variation on the landscape: Methods and cases. Annual Review of Ecology, Evolution and Systematics,43, 23–43.
Schulte, U., Hochkirch, A., Lötters, S., Rödder, D., Schweiger, S., Weimann, T., et al. (2012). Cryptic niche conservatism among evolutionary lineages of an invasive lizard. Global Ecology and Biogeography,21(2), 198–211.
Serra-Varela, M. J., Grivet, D., Vincenot, L., Broennimann, O., Gonzalo-Jiménez, J., & Zimmermann, N. E. (2015). Does phylogeographical structure relate to climatic niche divergence? A test using maritime pine (Pinus pinaster Ait.). Global Ecology and Biogeography,24(11), 1302–1313.
Smith, A. B., Godsoe, W., Rodríguez-Sánchez, F., Wang, H. H., & Warren, D. (2018). Niche estimation above and below the species level. Trends in Ecology and Evolution,34(3), 260–273.
Sokal, R. R., & Rohlf, F. J. (1995). Biometry (3rd ed.). New York: WH Freeman.
Sørensen, T. J. (1948). A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Copenhagen: I kommission hos E. Munksgaard.
Stewart, J. R. (2009). The evolutionary consequence of the individualistic response to climate change. Journal of Evolutionary Biology,22, 2363–2375.
Svetnik, V., Liaw, A., Tong, C., Culberson, J., Sheridan, R. P., & Feuston, B. P. (2003). Random forest: A classification and regression tool for compound classification and QSAR modeling. Journal of Chemical Information and Modeling,43, 1947–1958.
Tocchio, L. J., Gurgel-Gonçalves, R., Escobar, L. E., & Peterson, A. T. (2015). Niche similarities among white-eared opossums (Mammalia, Didelphidae): Is ecological niche modelling relevant to setting species limits? Zoologica Scripta,44(1), 1–10.
Valladares, F., Matesanz, S., Guilhaumon, F., Araújo, M. B., Balaguer, L., Benito-Garzón, M., et al. (2014). The effects of phenotypic plasticity and local adaptation on forecasts of species range shifts under climate change. Ecology Letters,17(11), 1351–1364.
Velásquez, N. A., Marambio, J., Brunetti, E., Méndez, M. A., Vásquez, R. A., & Penna, M. (2013). Bioacoustic and genetic divergence in a frog with a wide geographical distribution. Biological Journal of the Linnean Society,110(1), 142–155.
Victoriano, P., Ortiz, J. C., Troncoso, L., & Galleguillos, R. (1995). Allozyme variation in populations of Pleurodema thaul (Lesson, 1826) (Anura, Leptodactylidae). Comparative Biochemistry and Physiology B,112, 487–492.
Vidal, M. A., Soto, E. R., & Veloso, A. (2009). Biogeography of Chilean herpetofauna: distributional patterns of species richness and endemism. Amphibia-Reptilia,30, 151–171.
Walther, G. R., Post, E., Convey, P., Menzel, A., Parmesan, C., Beebee, T. J. C., et al. (2002). Ecological responses to recent climate change. Nature,416(6879), 389–395.
Wang, I. J. (2020). Topographic path analysis for modelling dispersal and functional connectivity: Calculating topographic distances using the topoDistance r package. Methods in Ecology and Evolution,11(2), 265–272.
Warren, D. L., Glor, R. E., & Turelli, M. (2008). Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution,62(11), 2868–2883.
Webb, C. O., Ackerly, D. D., McPeek, M. A., & Donoghue, M. J. (2002). Phylogenies and community ecology. Annual Review of Ecology, Evolution and Systematics,33, 475–505.
Wiens, J. J., & Graham, C. H. (2005). Niche conservatism: Integrating evolution, ecology, and conservation biology. Annual Review of Ecology, Evolution and Systematics,36, 519–539.
Williams, C. J. R. (2017). Climate change in Chile: An analysis of state-of-the-art observations, satellite-derived estimates and climate model simulations. Journal of Earth Science and Climatic Change,8(400), 2.
Acknowledgements
This study was funded through the Comisión Nacional de Investigación Científica y Tecnológica CONICYT Doctoral Fellowship (21120794) awarded to AMB. LDB acknowledges support from Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT Grant 1150029). SAE was funded by CONICYT PIA/BASAL FB0002. FAL acknowledges support from CONICYT (PIA ANILLOS Grant ACT172037) and from Universidad Santo Tomás (Grants TAS O00002256K and TAS O000022624). DZ, SAE and FAL acknowledge support from Fondo para la Innovación Agraria (Grant FIA-PYT-2016-0203).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Barria, A.M., Zamorano, D., Parada, A. et al. The Importance of Intraspecific Variation for Niche Differentiation and Species Distribution Models: The Ecologically Diverse Frog Pleurodema thaul as Study Case. Evol Biol 47, 206–219 (2020). https://doi.org/10.1007/s11692-020-09510-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-020-09510-0