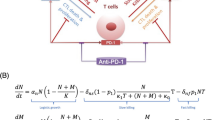
Abstract
LSD1 (KDMA1) has gained attention in the last decade as a cancer biomarker and drug target. In particular, recent work suggests that LSD1 inhibition alone reduces tumor growth, increases T cell tumor infiltration, and complements PD1/PDL1 checkpoint inhibitor therapy. In order to elucidate the immunogenic effects of LSD1 inhibition, we develop a mathematical model of tumor growth under the influence of the adaptive immune response. In particular, we investigate the anti-tumor cytotoxicity of LSD1-mediated T cell dynamics, in order to better understand the synergistic potential of LSD1 inhibition in combination immunotherapies, including checkpoint inhibitors. To that end, we formulate a non-spatial delay differential equation model and fit to the B16 mouse model data from Sheng et al. (Cell 174(3):549–563, 2018. https://doi.org/10.1016/j.cell.2018.05.052). Our results suggest that the immunogenic effect of LSD1 inhibition accelerates anti-tumor cytotoxicity. However, cytotoxicity does not seem to account for the slower growth observed in LSD1-inhibited tumors, despite evidence suggesting immune-mediation of this effect.











Similar content being viewed by others
Notes
We use the MCMC implementation from the R package BayesianTools, employing the differential evolution sampler ‘DEzs.’
References
Au-Yeung BB, Smith GA, Mueller JL, Heyn CS, Jaszczak RG, Weiss A, Zikherman J (2017) IL-2 modulates the TCR signaling threshold for CD8 but not CD4 T cell proliferation on a single-cell level. J Immunol 198(6):2445–2456. https://doi.org/10.4049/jimmunol.1601453
Augert A, Eastwood E, Ibrahim AH, Wu N, Grunblatt E, Basom R, Liggitt D, Eaton KD, Martins R, Poirier JT, Rudin CM, Milletti F, Cheng WY, Mack F, MacPherson D (2019) Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci Signal 12(567):eaau2922. https://doi.org/10.1126/scisignal.aau2922
Boulding T, McCuaig RD, Tan A, Hardy K, Wu F, Dunn J, Kalimutho M, Sutton CR, Forwood JK, Bert AG, Goodall GJ, Malik L, Yip D, Dahlstrom JE, Zafar A, Khanna KK, Rao S (2018) LSD1 activation promotes inducible EMT programs and modulates the tumour microenvironment in breast cancer. Sci Rep 8(1):73. https://doi.org/10.1038/s41598-017-17913-x
Chaudhary B, Elkord E (2016) Regulatory T cells in the tumor microenvironment and cancer progression: role and therapeutic targeting. https://doi.org/10.3390/vaccines4030028. www.mdpi.com/journal/vaccines
Chinen T, Kannan AK, Levine AG, Fan X, Klein U, Zheng Y, Gasteiger G, Feng Y, Fontenot JD, Rudensky AY (2016) An essential role for the IL-2 receptor in T reg cell function. Nat Immunol 17(11):1322–1333. https://doi.org/10.1038/ni.3540
Condamine T, Wang S, Diamond M, Hall L, Liu H, Chadderton A, Lu J, He C, Wu L, Burn T, Yao W, Hollis G, Huber R, Ruggeri B, Scherle P, Koblish H, Lee SH (2017) Abstract 4635: the LSD1 specific inhibitor INCB059872 enhances the activity of immune checkpoint blockade by reshaping the myeloid compartment in the syngeneic 4T1 mouse mammary tumor model. https://doi.org/10.1158/1538-7445.am2017-4635
Core UDB (2019) Differential gene expression analysis in R. https://ucdavis-bioinformatics-training.github.io/2019_March_UCSF_mRNAseq_Workshop/differential_expression/orig_DE_Analysis.html
De Boer RJ, Homann D, Perelson AS (2003) Different dynamics of CD4 + and CD8 + T cell responses during and after acute lymphocytic choriomeningitis virus infection. J Immunol. https://doi.org/10.4049/jimmunol.171.8.3928
De Pillis LG, Radunskaya AE, Wiseman CL (2005) A validated mathematical model of cell-mediated immune response to tumor growth. https://doi.org/10.1158/0008-5472.CAN-05-0564. www.aacrjournals.org
Eftimie R, Bramson JL, Earn DJD (2011) Interactions between the immune system and cancer: a brief review of non-spatial mathematical models. Bull Math Biol 73(1):2–32. https://doi.org/10.1007/s11538-010-9526-3
Eftimie R, Gillard JJ, Cantrell DA (2016) Mathematical models for immunology: current state of the art and future research directions. Bull Math Biol 78:2091–2134. https://doi.org/10.1007/s11538-016-0214-9
Feng Z, Yao Y, Zhou C, Chen F, Wu F, Wei L, Liu W, Dong S, Redell M, Mo Q, Song Y (2016) Pharmacological inhibition of LSD1 for the treatment of MLL-rearranged leukemia. J Hematol Oncol 9(1):24. https://doi.org/10.1186/s13045-016-0252-7
Gadhamsetty S, Marée AF, Beltman JB, de Boer RJ (2017) A sigmoid functional response emerges when Cytotoxic T lymphocytes start killing fresh target cells. Biophys J 112(6):1221–1235. https://doi.org/10.1016/j.bpj.2017.02.008
Gadhamsetty S, Marée AFM, Beltman JB, de Boer RJ (2014) A general functional response of Cytotoxic T lymphocyte-mediated killing of target cells. Biophys J 106(8):1780–1791. https://doi.org/10.1016/j.bpj.2014.01.048
Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, Miller CJ, Ogilvie DJ, Somervaille TC (2012) The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21(4):473–487. https://doi.org/10.1016/J.CCR.2012.03.014
Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, Nakamura Y, Hamamoto R (2011) Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer 128(3):574–586. https://doi.org/10.1002/ijc.25349
Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, Freeman GJ, Sharpe AH (2017) PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med 214(4):895–904. https://doi.org/10.1084/jem.20160801
Kaartinen T, Luostarinen A, Maliniemi P, Keto J, Arvas M, Belt H, Koponen J, Loskog A, Mustjoki S, Porkka K, Ylä-Herttuala S, Korhonen M (2017) Low interleukin-2 concentration favors generation of early memory T cells over effector phenotypes during chimeric antigen receptor T-cell expansion. Cytotherapy 19(6):689–702. https://doi.org/10.1016/j.jcyt.2017.03.067
Kalia V, Sarkar S (2018) Regulation of effector and memory CD8 T cell differentiation by IL-2-A balancing act. https://doi.org/10.3389/fimmu.2018.02987
Kashyap V, Ahmad S, Nilsson EM, Helczynski L, ead Kenna S, Liao Persson J, Gudas LJ, Mongan NP (2013) The lysine specific demethylase-1 (LSD1/KDM1A) regulates VEGF-A expression in prostate cancer. https://doi.org/10.1016/j.molonc.2013.01.003
Kavrochorianou N, Markogiannaki M, Haralambous S (2016) IFN-\(\beta \) differentially regulates the function of T cell subsets in MS and EAE. Cytokine Growth Factor Rev 30:47–54. https://doi.org/10.1016/j.cytogfr.2016.03.013
Kim PS, Lee PP, Levy D (2011) A theory of immunodominance and adaptive regulation. Bull Math Biol 73(7):1645–1665. https://doi.org/10.1007/s11538-010-9585-5
Kim PS, Lee PP, Levy D (2013) Basic principles in modeling adaptive regulation and immunodominance. Math Biosci 257:33–57. https://doi.org/10.1007/978-1-4614-4178-6_2
Kleppe M, Shank K, Efthymia P, Riehnhoff H, Levine RL (2015) Lysine-specific histone demethylase, LSD1, (KDM1A) as a novel therapeutic target in myeloproliferative neoplasms. Blood 126:23
Kuang Y, Nagy JD, Eikenberry SE (2018) Introduction to mathematical oncologyIntroduction to mathematical oncologyIntroduction to mathematical oncology. Chapman and Hall, London. https://doi.org/10.1201/9781315365404
Lechner MG, Karimi SS, Barry-Holson K, Angell TE, Murphy KA, Church CH, Ohlfest JR, Hu P, Epstein AL (2020) Immunogenicity of Murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. https://doi.org/10.1097/01.cji.0000436722.46675.4a
Lee W, Lee GR (2018) Transcriptional regulation and development of regulatory T cells. https://doi.org/10.1038/emm.2017.313
Li JL, Sainson RC, Shi W, Leek R, Harrington LS, Preusser M, Biswas S, Turley H, Heikamp E, Hainfellner JA, Harris AL (2007) Delta-like 4 notch Ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res 67(23):11244–11253. https://doi.org/10.1158/0008-5472.CAN-07-0969
Lim S, Janzer A, Becker A, Zimmer A, Schüle R, Buettner R, Kirfel J (2010) Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 31(3):512–520. https://doi.org/10.1093/carcin/bgp324
McKarns SC, Schwartz RH (2005) Distinct effects of TGF-\(\beta \)1 on CD4 + and CD8 + T cell survival, division, and IL-2 production: a role for T cell intrinsic Smad3. J Immunol 174(4):2071–2083. https://doi.org/10.4049/jimmunol.174.4.2071
McNally A, Hill GR, Sparwasser T, Thomas R, Steptoe RJ (2011) CD4+CD25+ regulatory T cells control CD8+ T-cell effector differentiation by modulating IL-2 homeostasis. Proc Natl Acad Sci USA 108(18):7529–7534. https://doi.org/10.1073/pnas.1103782108
Niebel D, Kirfel J, Janzen V, Höller T, Majores M, Gütgemann I (2014) Lysine-specific demethylase 1 (LSD1) in hematopoietic and lymphoid neoplasms. Blood 124(1):151–2. https://doi.org/10.1182/blood-2014-04-569525
Oh SA, Li MO (2013) TGF-\(\beta \): guardian of T cell function. J Immunol (Baltimore, Md.: 1950) 191(8):3973–3979. https://doi.org/10.4049/jimmunol.1301843
Potez M, Trappetti V, Bouchet A, Fernandez-Palomo C, Güç E, Kilarski WW, Hlushchuk R, Laissue J, Djonov V (2018) Characterization of a B16–F10 melanoma model locally implanted into the ear pinnae of C57BL/6 mice. PLoS ONE. https://doi.org/10.1371/journal.pone.0206693
Qin Y, Vasilatos SN, Chen L, Wu H, Cao Z, Fu Y, Huang M, Vlad AM, Lu B, Oesterreich S, Davidson NE, Huang Y (2018) Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene https://doi.org/10.1038/s41388-018-0451-5
Sakamoto A, Hino S, Nagaoka K, Anan K, Takase R, Matsumori H, Ojima H, Kanai Y, Arita K, Nakao M (2015) Lysine Demethylase LSD1 coordinates glycolytic and mitochondrial metabolism in hepatocellular carcinoma cells. Cancer Res 75(7):1445–1456. https://doi.org/10.1158/0008-5472.CAN-14-1560
Schenk T, Chen WC, Göllner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, Casero RA, Marton L, Woster P, Minden MD, Dugas M, Wang JCY, Dick JE, Müller-Tidow C, Petrie K, Zelent A, Zelent A (2012) Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med 18(4):605–11. https://doi.org/10.1038/nm.2661
Segarra M, Williams CK, De La M, Sierra L, Bernardo M, Mccormick PJ, Maric D, Regino C, Choyke P, Tosato G (2007) Dll4 activation of Notch signaling reduces tumor vascularity and inhibits tumor growth. https://doi.org/10.1182/blood-2007-11-126045. www.bloodjournal.org
Serce N, Gnatzy A, Steiner S, Lorenzen H, Kirfel J, Buettner R (2012) Elevated expression of LSD1 (Lysine-specific demethylase 1) during tumour progression from pre-invasive to invasive ductal carcinoma of the breast. BMC Clin Pathol 12:13. https://doi.org/10.1186/1472-6890-12-13
Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, Li Y, Chen H, Yang H, Hsu PH, Van Allen EM, Freeman GJ, De Carvalho DD, He HH, Sharpe AH, Shi Y (2018) LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174(3):549–563. https://doi.org/10.1016/j.cell.2018.05.052
Siekmann AF, Lawson ND, Siekmann AF, Lawson ND (2007) Extra view notch signalling and the regulation of angiogenesis addendum to: notch signalling limits angiogenic cell behavior in developing Zebrafish Arteries. Technical report 2. http://www.landesbioscience.com/journals/celladhesion/article/4488
Song M, Chen X, Wang L, Zhang Y (2018) Future of anti-PD-1/PD-L1 applications combinations with other therapeutic regimens. Chin J Cancer Res 30(2):157–172. https://doi.org/10.21147/j.issn.1000-9604.2018.02.01
Tammela T, Zarkada G, Wallgard E, Murtomäki A, Suchting S, Wirzenius M, Waltari M, Hellström M, Schomber T, Peltonen R, Freitas C, Duarte A, Isoniemi H, Laakkonen P, Christofori G, Ylä-Herttuala S, Shibuya M, Pytowski B, Eichmann A, Betsholtz C, Alitalo K (2008) Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature 454(7204):656–660. https://doi.org/10.1038/nature07083
Wodarz D, Komarova NL (2014) Dynamics of cancer. World Sci. https://doi.org/10.1142/8973
Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MYK, Duggan R, Wang Y, Barber GN, Fitzgerald KA, Alegre ML, Gajewski TF (2014) STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41(5):830–842. https://doi.org/10.1016/j.immuni.2014.10.017
Wyatt A (2019) Mathematical models of acute and chronic immunology. PhD thesis, University of Maryland
Yan Y, Kumar AB, Finnes H, Markovic SN, Park S, Dronca RS, Dong H (2018) Combining immune checkpoint inhibitors with conventional cancer therapy. Front Immunol 9:1739. https://doi.org/10.3389/fimmu.2018.01739
Yang GJ, Lei PM, Wong SY, Ma DL, Leung CH (2018) Pharmacological inhibition of LSD1 for cancer treatment. Molecules (Basel, Switzerland). https://doi.org/10.3390/molecules23123194
Ye C, Brand D, Zheng SG (2018) Targeting IL-2: an unexpected effect in treating immunological diseases. https://doi.org/10.1038/s41392-017-0002-5
Zafranskaya M, Oschmann P, Engel R, Weishaupt A, van Noort JM, Jomaa H, Eberl M (2007) Interferon-beta therapy reduces CD4+ and CD8+ T-cell reactivity in multiple sclerosis. Immunology 121(1):29–39. https://doi.org/10.1111/j.1365-2567.2006.02518.x
Zheng H, Jin B, Henrickson SE, Perelson AS, von Andrian UH, Chakraborty AK (2008) How antigen quantity and quality determine T-cell decisions in lymphoid tissue. Mol Cell Biol 28(12):4040–4051. https://doi.org/10.1128/MCB.00136-08
Acknowledgements
We would like to thank Heyrim Cho and Asia Wyatt for helpful discussions in drafting this paper. The work of JM was supported in part by the COMBINE Fellowship under NSF award DGE-1632976. The work of DL was supported in part by the National Science Foundation under Grant No. DMS-1713109.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Statistical Comparisons in Figures
For the pairwise comparisons in Figs. 4, 6, 8, and 9, we used an unpaired Student’s t test. For each panel, we applied a Benjamini–Hochberg adjustment for multiple testing. In order to validate the choice of a t test, we used the Shapiro–Wilk test for normality. We cannot reject normality for the samples in Figs. 4, 6, and 8 (\(P > 0.05\)). For Fig. 9, two of the samples were somewhat non-normal. Left to right in Fig. 9, the S–W statistic had values 0.93, 0.85, 0.90, 0.98 corresponding to \(P=0.17, 8.8e{-}4, 0.01, 0.81\). We used the same comparison for Fig. 4d as in Sheng et al. (2018).
For the differential gene expression (DGE) analysis for Fig. 7, we used the gene counts from Sheng et al. (2018) (data accessible at NCBI GEO database, accession GSE112230) and the R package edgeR. We normalized the counts using the TMM method and removed minimally expressing genes, leaving us with 12,305 genes remaining. We fit a linear model to compare gene expression between the three experimental tumor conditions: scramble control, LSD1-KO, and LSD1/MDA5 DKO tumors. Our workflow was based upon the tutorial in Core (2019). When adjusting for multiple testing, we used the BH method as before and adjusted for all three pairwise contrasts (between our experimental conditions) for the full set of expressing genes, together.
Description of Statistical Model and MCMC Fitting
Consider the tumor growth data for tumor i as a time series \({\varvec{y}}_i = \left( y_i(t_j) \right) _{j=1}^{n_i}\). We assume a statistical model of the form
where \(f({\varvec{p}}_i, \cdot )\) is a deterministic model and \(\epsilon _i(\cdot )\) is the measurement noise, parameterized by \({\varvec{p}}_i, \mu _i, \sigma _i\) individually for each tumor time series i. The model \(f({\varvec{p}}_i, \cdot )\) is the solution to either our main DDE model (1) described in Sect. 2.1, (1), or one of the alternative ODE models in Sect. 2.1 (2, 3, 4). We use \({\varvec{f}}({\varvec{p}}_i)\) to denote \( \left( f({\varvec{p}}_i, t_j) \right) _j\), i.e., the estimated time series from our model, corresponding to the data \({\varvec{y}}_i\). For our measurement noise, we ideally would have \(\mu _i = 0\) for each tumor. We make this assumption for model fitting, but estimate \(\mu _i\) for the purposes of model validation below, in order to strengthen the likelihood of simple alternative models.
Given a model f and parameters \({\varvec{p}}_i, \mu _i, \sigma _i\), the conditional log-likelihood is given by
For the purposes of model fitting, we estimated \({\hat{{\varvec{p}}}}_i\) for fixed \(\sigma _i\) using a Markov chain Monte Carlo,Footnote 1 under the assumption that \(\mu _i = 0\). We linearly interpolated our data \({\varvec{y_i}} \mapsto {\tilde{{\varvec{y}}}_i}\) so that we had 5 data points per day, in order to ensure smooth fits. Per standard practice, we employ the \(\ell _2\) error as the target function g, for which \(e^g \propto L\):
To find \({\varvec{f}}({\varvec{p}}_i)\), we need to solve systems (1, 2, 3, 4). The one-dimensional systems 2, 3 have well-known closed-form solutions:
For our main model (1) and the two compartment model (4), we solved our equations numerically using the R package diffeqr, which is a convenient wrapper for the Julia suite DifferentialEquations.jl.
To validate our model, we compared it to each of the alternative choices of f, for each tumor i, using the standard Bayesian Information Criterion (BIC):
where \(N_p\) is the number of free parameters for our model, i.e., the length of \(({\varvec{p}}_i, \sigma _i)\). In particular, these are 7, 4, 4, 6 for models (1), (2), (3), and (4), respectively. Substituting (7) into (11) and using our estimate \({\varvec{p}}_i\), we have
Supplementary Tables
Marginal Parameter Densities
See Fig. 12.
Prior and posterior parameter distribution for main model fitting (LSD1-KO #5). Presented are the prior and posterior distribution of the parameters \({\varvec{p}}_i\) of model (1), taken from the MCMC, where i is one of the LSD1-KO tumors. In order from left to right, the panels here correspond to \(\alpha , \mu , \ell , r, \log _{10}(s_{\mathrm {H}}), \log _{10}(s_K)\)
Rights and permissions
About this article
Cite this article
Milzman, J., Sheng, W. & Levy, D. Modeling LSD1-Mediated Tumor Stagnation. Bull Math Biol 83, 15 (2021). https://doi.org/10.1007/s11538-020-00842-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-020-00842-8