Abstract
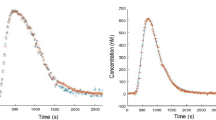
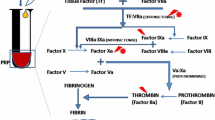
The coagulation cascade comprises numerous chemical reactions between many proteins, that finally lead to the formation of a clot to stop bleeding. Many numerical models have attempted to translate understanding of this cascade into mathematical equations that simulate the chain reactions. However, their predictions have not been validated against clinical data stemming from patients. In this paper, we propose an extensive validation of five available models, by comparing in healthy and haemophilic subjects, thrombin generation measured in vitro to thrombin generation predicted by the models in silico. In order to render the models more predictive, we calibrated the models to have an acceptable agreement between the experimental and estimated data. Optimization processes based on genetic algorithms were developed to search for those calibrated kinetic parameters. Our results show that the thrombin generation kinetics are so complex that they cannot be predicted by a unique set of kinetic parameters for all patients: the calibration of only three parameters in a subject-specific way allows reaching good model estimations for different experimental conditions realized on the same patient.
















Similar content being viewed by others
Abbreviations
- CAT:
-
Calibrated automated thrombogram
- CTI:
-
Corn trypsin inhibitor
- CV:
-
Coefficient of variation
- ETP:
-
Endogenous thrombin potential
- GA:
-
Genetic algorithm
- HA:
-
Haemophilia A
- HB:
-
Haemophilia B
- LT:
-
Lag time
- M:
-
\(\hbox {mol.\,L}^{-1}\)
- ODE:
-
Ordinary differential equation
- PPP:
-
Platelet-poor plasma
- ST:
-
Start tail
- TG:
-
Thrombin generation
- ttP:
-
Time to peak
- II:
-
Prothrombin
- IIa:
-
Thrombin
- V:
-
Factor V
- Va:
-
Activated factor V
- VII:
-
Factor VII
- VIIa:
-
Activated factor VII
- VIII:
-
Factor VIII
- VIIIa:
-
Activated factor VIII (anti-haemophilic A factor)
- IX:
-
Factor IX
- IXa:
-
Activated factor IX (anti-haemophilic B factor)
- X:
-
Factor X
- Xa:
-
Activated factor X
- XI:
-
Factor XI
- XIa:
-
Activated factor XI
- XII:
-
Factor XII
- XIIa:
-
Activated factor XII
- AT:
-
Antithrombin
- Fbg:
-
Fibrinogen
- Fbn:
-
Fibrin
- K:
-
Kallikrein
- PC:
-
Protein C
- PK:
-
Prekallikrein
- PS:
-
Protein S
- TF:
-
Tissue factor
- TFPI:
-
Tissue factor pathway inhibitor
- TM:
-
Thrombomodulin
- \(\mathbf{C }\) :
-
Vector of the concentrations of the chemical species in the plasma
- e :
-
Relative error between model predictions and experimental data
- i :
-
Subject index
- k :
-
Kinetic parameter
- \({\tilde{k}}\) :
-
Normalized kinetic parameter
- \(\mathbf{k }\) :
-
Vector of kinetic parameters
- \(\tilde{\mathbf{k }}\) :
-
Vector of normalized kinetic parameters
- \(L_1\) :
-
Cost function used to search for optimized sets of kinetic parameters
- \(L^{exp}_{i,l}\) :
-
Variance of the experimental TG curve of subject i in the l-th experimental condition
- M :
-
Number of discrete timepoints at which TG was measured
- n :
-
Number of parameters characterizing the TG curves
- N :
-
Number of subjects in a subgroup of the study population
- \(N_\mathrm{p}\) :
-
Number of kinetic parameters in a model
- \(N_\mathrm{reactions}\) :
-
Number of reactions in a model
- \(N_\mathrm{s}\) :
-
Number of subjects included in the study
- \(N_\mathrm{var}\) :
-
Number of variables in a model
- \(p_0\) :
-
Weight parameter
- r :
-
Function of the change in concentration of a chemical species
- \(\mathbf{R }\) :
-
Vector-valued function of the changes in concentrations of all the chemical species introduced in a model
- \(R^2\) :
-
Coefficient of determination
- \(t_m\) :
-
Discrete time instants over which the TG assays run
- \(\epsilon \) :
-
Set of experimental conditions
- \(\sigma \) :
-
Standard deviation of the normalized kinetic parameter \({\tilde{k}}\) over all sets of kinetic parameters in \({\varOmega }\)
- \({\varOmega }\) :
-
Set of vectors of kinetic parameters providing a good accuracy
- [a]:
-
Concentration of protein a
- |a|:
-
Cardinality of set a
- \({\bar{a}}\) :
-
Average of the variable a
- \({\tilde{a}}\) :
-
Normalized value of a
- \(_i\) :
-
... related to subject i
- \(_j\) :
-
... related to the parameter \(p_j\) of the TG curve
- \(_l\) :
-
... related to the lth experimental condition
- \(_m\) :
-
... related to the timepoint m of the TG curve
- \(^0\) :
-
... at time \({t}=0\)
- \(^\mathrm{calib}\) :
-
... used for the calibration process
- \(^\mathrm{exp}\) :
-
... determined experimentally
- \(^\mathrm{model}\) :
-
... as introduced in the original model
- \(^{p}\) :
-
... related to the parameter p of the TG curve
- \(^\mathrm{pop}\) :
-
... uniform over a subgroup of subjects
- \(^\mathrm{pred}\) :
-
... predicted by the model
- \(^\mathrm{valid}\) :
-
... used for the validation process
References
Abgrall JF (1995) Physiologie de la coagulation. Hematoweb, Paris
Anand M, Rajagopal K, Rajagopal KR (2003) A model incorporating some of the mechanical and biochemical factors underlying clot formation and dissolution in flowing blood. J Theor Med 5(3–4):183–218. https://doi.org/10.1080/10273660412331317415
Baugh RJ (1998) Regulation of extrinsic pathway factor Xa formation by tissue factor pathway inhibitor. J Biol Chem 273(8):4378–4386. https://doi.org/10.1074/jbc.273.8.4378
Biasetti J, Spazzini PG, Swedenborg J, Gasser TC (2012) An integrated fluid-chemical model toward modeling the formation of intra-luminal thrombus in abdominal aortic aneurysms. Frontiers Physiol 3:266. https://doi.org/10.3389/fphys.2012.00266
Bodnar T, Sequeira A, Prosi M (2011) On the shear-thinning and viscoelastic effects of blood flow under various flow rates. Appl Math Comput 217(11):5055–5067. https://doi.org/10.1016/j.amc.2010.07.054
Li Braescu, George T, Orbulescu C, Leretter M (2009) Dynamics of thrombin formation using a mathematical model including both intrinsic and extrinsic pathways of blood coagulation. Dyn Contin Discret I (Series A) 16(S1):75–82
Brummel-Ziedins K, Orfeo T, Jenny N, Everse S, Mann K (2003) Wintrobe’s Clinical Hematology, Lippincott Williams & Wilkins, Philidelphia, chap Blood coagulation and fibrinolysis, pp 667–774
Bungay S (2008) Modelling the effect of amplification pathway factors on thrombin generation: a comparison of hemophilias. Transfus Apheresis Sci 38(1):41–47. https://doi.org/10.1016/j.transci.2007.12.007
Bungay S, Gentry P, Gentry R (2003) A mathematical model of lipid-mediated thrombin generation. Math Med Biol 20(1):105–129
Chatterjee M, Denney WS, Jing H, Diamond SL (2010) Systems biology of coagulation initiation: kinetics of thrombin generation in resting and activated human blood. PLOS Comput Biol 6(9):e1000950. https://doi.org/10.1371/journal.pcbi.1000950
Danforth CM, Orfeo T, Mann KG, Brummel-Ziedins KE, Everse SJ (2009) The impact of uncertainty in a blood coagulation model. Math Med Biol 26(4):323–336. https://doi.org/10.1093/imammb/dqp011
Dielis A, Castoldi E, Spronk HMH, Van Oerle R, Hamulyak K, Ten Cate H, Rosing J (2008) Coagulation factors and the protein C system as determinants of thrombin generation in a normal population. J Thromb Haemost 6(1):125–131. https://doi.org/10.1111/j.1538-7836.2007.02824.x
Engl HW, Flamm C, Kügler P, Lu J, Müller S, Schuster P (2009) Inverse problems in systems biology. Inverse Prob 25(12):123,014. https://doi.org/10.1088/0266-5611/25/12/123014
Fay PJ, Haidaris PJ, Smudzin TM (1991) Human factor VIIIa subunit structure. Reconstruction of factor VIIIa from the isolated A1/A3-C1-C2 dimer and A2 subunit. J Biol Chem 266(14):8957–8962
Fedosov DA, Caswell B, Karniadakis GE (2010) A multiscale red blood cell model with accurate mechanics, rheology, and dynamics. Biophys J 98(10):2215–2225. https://doi.org/10.1016/j.bpj.2010.02.002
Fogelson AL, Guy RD (2008) Immersed-boundary-type models of intravascular platelet aggregation. Comput Methods Appl Mech Eng 197(25–28):2087–2104. https://doi.org/10.1016/j.cma.2007.06.030
Goldberg DE (1989) Genetic algorithms in search, optimization and machine learning. Addison-Wesley, Boston
Hemker HC, Béguin S (2000) Phenotyping the clotting system. Thromb Haemost 84(5):747–751
Hemker HC, Giesen P, AlDieri R, Regnault V, De Smed E, Wagenvoord R, Lecompte T, Béguin S (2003) The calibrated automated thrombogram (CAT): a universal routine test for hyper-and hypocoagulability. Pathophysiol Haemost Thromb 32(5–6):249–253
Hockin MF (2002) A model for the stoichiometric regulation of blood coagulation. J Biol Chem 277(21):18,322–18,333. https://doi.org/10.1074/jbc.M201173200
Hoffmeister F, Bäck T (1991) Genetic algorithms and evolution strategies: similarities and differences. In: Schwefel HP, Männer R (eds) Parallel problem solving from nature. Springer, Heidelberg, pp 455–469
Hund SJ (2010) A computational model of platelet mediated thrombosis for the evaluation and design of medical devices. PhD thesis, Biomedical Engineering, Carnegie Mellon University
Jaqaman K, Danuser G (2006) Linking data to models: data regression. Nat Rev Mol Cell Biol 7:813–819
Jones KC, Mann KG (1994) A model for the tissue factor pathway to thrombin. J Biol Chem 269(37):23,367–23,378
Jordan S, Chaikof E (2011) Simulated surface-induced thrombin generation in a flow field. Biophys J 101(2):276–286. https://doi.org/10.1016/j.bpj.2011.05.056
Khanin MA, Rakov DV, Kogan AE (1998) Mathematical model for the blood coagulation prothrombin time test. Thromb Res 89(5):227–232
Kuharsky AL, Fogelson AL (2001) Surface-mediated control of blood coagulation: the role of binding site densities and platelet deposition. Biophys J 80(3):1050–1074
LaCroix D (2012) A reduced equation mathematical model for blood coagulation and fibrinolysis in quiescent plasma. Int J Struct Changes Solids 4:23–35
Lagarias JC, Reeds JA, Wright MH, Wright PE (1998) Convergence properties of the Nelder–Mead simplex method in low dimensions. SIAM J Optim 9(1):112–147
Lollar P, Parker ET, Fay PJ (1992) Coagulant properties of hybrid human/porcine factor VIII molecules. J Biol Chem 267(33):23652–23657
Luan D, Zai M, Varner JD (2007) Computationally derived points of fragility of a human cascade are consistent with current therapeutic strategies. PLoS Comput Biol 3(7):e142. https://doi.org/10.1371/journal.pcbi.0030142
Makin JG, Narayanan S (2013) A hybrid-system model of the coagulation cascade: simulation, sensitivity, and validation. J Bioinform Comput Biol 11(5):1342004. https://doi.org/10.1142/S0219720013420043
Mann KG (2003) The dynamics of thrombin formation. Arterioscler Thromb Vasc Biol 23(1):17–25. https://doi.org/10.1161/01.ATV.0000046238.23903.FC
Margaria G, Riccomagno E, Chappell MJ, Wynn HP (2001) Differential algebra methods for the study of the structural identifiability of rational function state-space models in the biosciences. Math Biosci 174(1):1–26. https://doi.org/10.1016/S0025-5564(01)00079-7
Moles CG, Mendes P, Banga JR (2003) Parameter estimation in biochemical pathways: a comparison of global optimization methods. Genome Res 13(11):2467–2474
Morrissey J, Gail Macik B, Neuenschwander P, Comp P (1993) Quantitation of activated factor vii levels in plasma using a tissue factor mutant selectively deficient in promoting factor vii activation. Blood 81(3):734–744
Panteleev MA, Ovanesov MV, Kireev DA, Shibeko AM, Sinauridze EI, Ananyeva NM, Butylin AA, Saenko EL, Ataullakhanov FI (2006) Spatial propagation and localization of blood coagulation are regulated by intrinsic and protein C pathways, respectively. Biophys J 90(5):1489–1500. https://doi.org/10.1529/biophysj.105.069062
Panteleev MA, Balandina AN, Lipets EN, Ovanesov MV, Ataullakhanov FI (2010) Task-oriented modular decomposition of biological networks: trigger mechanism in blood coagulation. Biophys J 98(9):1751–1761. https://doi.org/10.1016/j.bpj.2010.01.027
Polisetty PK, Voit EO, Gatzke EP (2006) Identification of metabolic system parameters using global optimization methods. Theor Biol Med Model 3(1):4
Qiao Y, Xu C, Zeng Y, Xu X, Zhao H, Xu H (2004) The kinetic model and simulation of blood coagulation—the kinetic influence of activated protein C. Med Eng Phys 26(4):341–347. https://doi.org/10.1016/j.medengphy.2004.01.003
Qiao Y, Liu J, Zeng Y (2005) A kinetic model for simulation of blood coagulation and inhibition in the intrinsic path. J Med Eng Technol 29(2):70–74. https://doi.org/10.1080/03091900410001709079
Rittenhouse MA, Revie CW, Hurford A (2016) A model for sea lice (Lepeophtheirus salmonis) dynamics in a seasonally changing environment. Epidemics 16:8–16
Rodriguez-Fernandez M, Egea J, Banga J (2006a) Novel metaheuristic for parameter estimation in nonlinear dynamic biological systems. BMC Bioinform 7:483. https://doi.org/10.1186/1471-2105-7-483
Rodriguez-Fernandez M, Mendes P, Banga JR (2006b) A hybrid approach for efficient and robust parameter estimation in biochemical pathways. Biosystems 83(2):248–265. https://doi.org/10.1016/j.biosystems.2005.06.016 (5th International Conference on Systems Biology)
Rodriguez-Fernandez M, Rehberg M, Kremling A, Banga JR (2013) Simultaneous model discrimination and parameter estimation in dynamic models of cellular systems. BMC Syst Biol 7(1):76. https://doi.org/10.1186/1752-0509-7-76
Rosing J, Tans G, Govers-Riemslag J, Zwaal R, Hemker H (1980) The role of phospholipids and factor Va in the prothrombinase complex. J Biol Chem 255(1):274–283
Runarsson TP, Yao X (2005) Search biases in constrained evolutionary optimization. IEEE Trans Syst Man Cybern Part C Appl Rev 35(2):233–243. https://doi.org/10.1109/TSMCC.2004.841906
Saravanan N, Fogel DB, Nelson KM (1995) A comparison of methods for self-adaptation in evolutionary algorithms. Biosystems 36(2):157–166. https://doi.org/10.1016/0303-2647(95)01534-R
Stonebraker J, Bolton-Maggs P, Soucie J, Walkers I, Brooker M (2010) A study of variations in the reported haemophilia a prevalence around the world. Haemophilia 16:20–32. https://doi.org/10.1111/j.1365-2516.2009.02127.x
Stonebraker J, Bolton-Maggs P, Soucie J, Walkers I, Brooker M (2011) A study of variations in the reported haemophilia b prevalence around the world. Haemophilia 16(1):20–32. https://doi.org/10.1111/j.1365-2516.2011.02588.x
Subcommittee on Control of Anticoagulation of the SSC of the ISTH (2011) Towards a recommendation for the standardization of the measurement of platelet-dependent thrombin generation. J Thromb Haemost 9:1859–1861. https://doi.org/10.1111/j.1538-7836.2011.04427.x
Tosenberger A, Ataullakhanov F, Bessonov N, Panteleev M, Tokarev A, Volpert V (2013) Modelling of thrombus growth in flow with a DPD-PDE method. J Theor Biol 337:30–41. https://doi.org/10.1016/j.jtbi.2013.07.023
Tracy P, LL E, Mann K (1985) Human prothrombinase complex assembly and function on isolated peripheral blood cell populations. J Biol Chem 260(4):2119–2124
Tyurin KV, Khanin MA (2006) Hemostasis as an optimal system. Math Biosci 204(2):167–184. https://doi.org/10.1016/j.mbs.2006.09.005
Wajima T, Isbister GK, Duffull SB (2009) A comprehensive model for the humoral coagulation network in humans. Clin Pharmacol Therap 86(3):290–298. https://doi.org/10.1038/clpt.2009.87
White LJ, Evans ND, Lam TJGM, Schukken YH, Medley GF, Godfrey KR, Chappell MJ (2002) The structural identifiability and parameter estimation of a multispecies model for the transmission of mastitis in dairy cows with postmilking teat disinfection. Math Biosci 180:275–91
Xu CQ, Zeng YJ, Gregersen H (2002) Dynamic model of the role of platelets in the blood coagulation system. Med Eng Phys 24(9):587–593
Xu C, Hu XuX, Zeng Y, Wen Chen Y (2005) Simulation of a mathematical model of the role of the TFPI in the extrinsic pathway of coagulation. Comput Biol Med 35(5):435–445. https://doi.org/10.1016/j.compbiomed.2004.02.007
Xu Z, Lioi J, Mu J, Kamocka MM, Liu X, Chen DZ, Rosen ED, Alber M (2010) A multiscale model of venous thrombus formation with surface-mediated control of blood coagulation cascade. Biophys J 98(9):1723–1732. https://doi.org/10.1016/j.bpj.2009.12.4331
Zarnitsina VI, Pokhilko AV, Ataullakhanov FI (1996a) A mathematical model for the spatio-temporal dynamics of intrinsic pathway of blood coagulation. I. The model description. Thromb Res 84(4):225–236
Zarnitsina VI, Pokhilko AV, Ataullakhanov FI (1996b) A mathematical model for the spatio-temporal dynamics of intrinsic pathway of blood coagulation. II. Results. Thromb Res 84(5):333–344
Zhu D (2007) Mathematical modeling of blood coagulation cascade: kinetics of intrinsic and extrinsic pathways in normal and deficient conditions. Blood Coagul Fibrinolysis 18(7):637–646
Zhu A, Guo J, Ni BJ, Wang S, Yang Q, Peng Y (2015) A novel protocol for model calibration in biological wastewater treatment. Sci Rep 5:8493. https://doi.org/10.1038/srep08493
Acknowledgements
The authors are grateful to Pfizer for its financial support for the experimental procedures and to Diagnostica Stago for its material support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chelle, P., Morin, C., Montmartin, A. et al. Evaluation and Calibration of In Silico Models of Thrombin Generation Using Experimental Data from Healthy and Haemophilic Subjects. Bull Math Biol 80, 1989–2025 (2018). https://doi.org/10.1007/s11538-018-0440-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-018-0440-4