Abstract
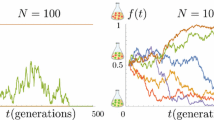
Genetic diversity grows with the population size in most neutral evolution models. Empirical evidence of large populations with limited diversity has been proposed to be due either to genetic bottlenecks or to selection. An alternative explanation is that the limited diversity is a result of rare reproduction events. Indeed, recent estimates of the offspring number distribution highlight the role of large reproduction events. We here show that in a large class of models containing such rare events, genetic diversity decreases as the population size increases, in neutral evolution models. For many realistic offspring number distributions, the contribution of rare events to the dynamics grows with the population size. In the context of genetic diversity, these rare events induce a decrease in the time to the most recent common ancestor and in the genetic heterogeneity as the population grows. This phenomenon may explain the observed rapid fixation of genes in large populations, in the absence of observable selection or bottlenecks.





Similar content being viewed by others
References
Arnason E (2004) Mitochondrial cytochrome b DNA variation in the high-fecundity Atlantic cod: trans-Atlantic clines and shallow gene genealogy. Genetics 166:1871–1885
Athreya K (2012) Coalescence in the recent past in rapidly growing populations. Stoch Process Appl 122:3757–3766
Eldon B, Wakeley J (2006) Coalescent processes when the distribution of offspring number among individuals is highly skewed. Genetics 172:2621–2633
Ewens WJ (2004) Mathematical population genetics 1: I. Theoretical introduction. Springer, Berlin
Fields B et al (2007) Fields’ virology, 5th edn. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia
Haas M, Pigorsch C (2009) Financial economics, fat-tailed distributions. Encycl Complex Syst Sci 4:3404–3435
Hedgecock D, Pudovkin AI (2011) Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull Mar Sci 87:971–1002
Hedgecock D, Tracey M, Nelson K (1982) Genetics, the biology of crustacea. Academic Press, New York
Hoban SM, Mezzavilla M, Gaggiotti OE, Benazzo A, Van Oosterhout C, Bertorelle G (2013) High variance in reproductive success generates a false signature of a genetic bottleneck in populations of constant size: a simulation study. BMC Bioinformatics 14(1):1–10
Huillet T, Möhle M (2013) On the extended Moran model and its relation to coalescents with multiple collisions. Theor Popul Biol 87:5–14
Kingman JF (1982a) On the genealogy of large populations. J Appl Probab 27–43
Kingman JFC (1982b) The coalescent. Stoch Process Appl 13:235–248
Lambert A (2008) Population dynamics and random genealogies. Stoch Models 24:45–163
Leonarduzzi C et al (2012) Towards an optimal sampling effort for paternity analysis in forest trees: what do the raw numbers tell us? iFor-Biogeosci For 5:18–25
Möhle M, Sagitov S (2001) A classification of coalescent processes for haploid exchangeable population models. Ann Probab 29:1547–1562
Nei M et al (1975) The bottleneck effect and genetic variability in populations. Evolution 29(1):1–10
Pitman J (1999) Coalescents with multiple collisions. Ann Probab 27(4):1870–1902
Resnick SI (2007) Heavy-tail phenomena: probabilistic and statistical modeling. Springer, Berlin
Schweinsberg J (2003) Coalescent processes obtained from supercritical Galton–Watson processes. Stoch Proc Appl 106(1):107–139
Turner TF et al (1999) Temporal genetic variation of mitochondrial DNA and the female effective population size of red drum (Sciaenops ocellatus) in the northern Gulf of Mexico. Molec Ecol 8:1223–1229
Wakeley J (2009) Coalescent theory: an introduction. Roberts & Company Publishers, Englewood
Wakeley J, Sargsyan O (2009) Extensions of the coalescent effective population size. Genetics 181:341–345
Zerjal T et al (2003) The genetic legacy of the Mongols. Am J Hum Genet 72:717–721
Acknowledgments
We thank Gur Yaari, Reuven Cohen, Naama Brenner and Marc Feldman for their comments and Miriam Beller for the English editing.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1: Asymptotic Analysis of Truncated Power Laws
Let \(U_N\) be a r.v. with truncated power law distribution:
where \(D_N\) is the normalizing constant. Note that
The asymptotic approximations of these sums are well known (we write \(f\sim g\) for \(\lim \limits _{N\rightarrow \infty } f/g=1)\):
where \(\zeta \) denotes the Riemann zeta function. From this, it immediately follows that
and
Thus, we can conclude that in the notation of Theorem 1, the choice of MC as a modified Moran model with \(U_N\) distributed as above will lead to MG having a decreasing TMRCA of a sample as time passes if and only if \(\alpha <2\).
Appendix 2: Simulation Methods
For the growing population model, we simulate the entire process forward in time. First we generate the times of reproduction events as exponentials with rate equal to N, the current population size. Then we generate the embedded jump chain by drawing at each transition a (uniformly) random member of the population to reproduce, drawing his number of offspring from the law of \(U_N +c\) and drawing a random sample of size \(N-U_N\) from the previous generation who will survive to the next generation. We iterate this (with different values of N), maintain the entire history of the embedded chain in memory and average out the waiting times to recover a realization of the continuous process.
To calculate the TMRCA of a sample, we simply follow its ancestral line in the generated lineage tree up to the first occurrence of a single ancestor.
To calculate the heterozigosity and mean pairwise difference of the population, we superimposed a mutation model independent of reproduction events, with exponentially distributed mutation events.
Rights and permissions
About this article
Cite this article
Alter, I., Louzoun, Y. Population Growth Combined with Wide Offspring Distributions can Increase Fixation Rate and Reduce Genetic Diversity. Bull Math Biol 78, 1477–1492 (2016). https://doi.org/10.1007/s11538-016-0191-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-016-0191-z