Abstract
The earliest cell fate decisions in a developing embryo are those associated with establishing the germ layers. The specification of the mesoderm and endoderm is of particular interest as the mesoderm is induced from the endoderm, potentially from an underlying bipotential group of cells, the mesendoderm. Mesendoderm formation has been well studied in an amphibian model frog, Xenopus laevis, and its formation is driven by a gene regulatory network (GRN) induced by maternal factors deposited in the egg. We have recently demonstrated that the axolotl, a urodele amphibian, utilises a different topology in its GRN to specify the mesendoderm. In this paper, we develop spatially structured mathematical models of the GRNs governing mesendoderm formation in a line of cells. We explore several versions of the model of mesendoderm formation in both Xenopus and the axolotl, incorporating the key differences between these two systems. Model simulations are able to reproduce known experimental data, such as Nodal expression domains in Xenopus, and also make predictions about how the positional information derived from maternal factors may be interpreted to drive cell fate decisions. We find that whilst cell–cell signalling plays a minor role in Xenopus, it is crucial for correct patterning domains in axolotl.






Similar content being viewed by others
References
Alon U (2007) An introduction to systems biology: design principles of biological circuits. Chapman and Hall/CRC, Boca Raton
Brown L, King J, Loose M (2014) Two different network topologies yield bistability in models of mesoderm and anterior mesendoderm specification in amphibians. J Theor Biol 353:67–77
Cha Y, Takahashi S, Wright C (2006) Cooperative non-cell and cell autonomous regulation of Nodal gene expression and signaling by Lefty/Antivin and Brachyury in Xenopus. Dev Biol 290(2):246–264
Chen Y (2010) Mesoderm induction in ambystoma mexicanum, a urodele amphibian. PhD thesis, University of Nottingham
Chisholm R, Hughes B, Landman K (2010) Building a morphogen gradient without diffusion in a growing tissue. PLoS One 5(9):e12,857. doi:10.1371/journal.pone.0012857
Fáth G (1998) Propagation failure of traveling waves in a discrete bistable medium. Phys D 116:176–190
Gilbert S (2010) Developmental biology, 9th edn. Sinauer Associates, Sunderland
Green J (2002) Morphogen gradients, positional information, and Xenopus: interplay of theory and experiment. Dev Dyn 225(4):392–408
Green J, Smith J (1990) Graded changes in dose of a Xenopus activin A homologue elicit stepwise transitions in embryonic cell fate. Nature 347:391–394
Gurdon J, Harger P, Mitchell A, Lemaire P (1994) Activin signalling and response to a morphogen gradient. Nature 371:487–492
Gurdon J, Mitchell A, Ryan K (1996) An experimental system for analyzing response to a morphogen gradient. Proc Natl Acad Sci USA 93(18):9334–9338
Gurdon J, Standley H, Dyson S, Butler K, Langon T, Ryan K, Stennard F, Shimizu K, Zorn A (1999) Single cells can sense their position in a morphogen gradient. Development 126(23):5309–5317
Hansen C, Marion C, Steele K, George S, Smith W (1997) Direct neural induction and selective inhibition of mesoderm and epidermis inducers by Xnr3. Development 124(2):483–492
Heasman J (2006) Maternal determinants of embryonic cell fate. Seminn Cell Dev Biol 17(1):93–98. doi:10.1016/j.semcdb.2005.11.005
Jones C, Armes N, Smith J (1996) Signalling by TGF-beta family members: short-range effects of Xnr-2 and BMP-4 contrast with the long-range effects of Activin. Curr Biol 6:1468–1475
Keener J (1987) Propagation and its failure in coupled systems of discrete excitable cells. SIAM J Appl Math 47(3):556–572
Keener J, Sneyd J (2009) Mathematical physiology: cellular physiology, vol 1. Springer, New York
Kerszberg M, Wolpert L (1998) Mechanisms for positional signalling by morphogen transport: a theoretical study. J Theor Biol 191(1):103–114. doi:10.1006/jtbi.1997.0575
Kimelman D (2006) Mesoderm induction: from caps to chips. Nat Rev Genet 7(5):360–72
Lander A, Nie Q, Wan F (2006) Internalization and end flux in morphogen gradient formation. J Comput Appl Math 190(1–2):232–251. doi:10.1016/j.cam.2004.11.054
Lee M, Heasman J, Whitman M (2001) Timing of endogenous activin-like signals and regional specification of the Xenopus embryo. Development 128(15):2939–2952
Lemaire P (1998) A role for the vegetally expressed Xenopus gene Mix. 1 in endoderm formation and in the restriction of mesoderm to the marginal zone. Development 125:2371–2380
Lerchner W, Latinkic B, Remacle J, Huylebroeck D, Smith J (2000) Region-specific activation of the xenopus brachyury promoter involves active repression in ectoderm and endoderm: a study using transgenic frog embryos. Development 127(12):2729–2739
Luxardi G, Marchal L, Thomé V, Kodjabachian L (2010) Distinct Xenopus Nodal ligands sequentially induce mesendoderm and control gastrulation movements in parallel to the Wnt/PCP pathway. Development 137(3):417–426
Middleton A (2007) Mathematical modelling of gene regulatory networks. PhD thesis
Middleton A, King J, Loose M (2009) Bistability in a model of mesoderm and anterior mesendoderm specification in Xenopus laevis. J Theor Biol 260(1):41–55
Middleton A, King J, Loose M (2013) Wave pinning and spatial patterning in a mathematical model of Antivin/LeftyNodal signalling. J Math Biol 67(6–7):1393–1424
Muratov C, Shvartsman S (2004) Signal propagation and failure in discrete autocrine relays. Phys Rev Lett 93:118101
Nahmad M, Lander A (2011) Spatiotemporal mechanisms of morphogen gradient interpretation. Curr Opin Genet Dev 21(6):726–731
Nath K, Ellison R (2007) RNA of AxVegT, the axolotl orthologue of the Xenopus meso-endoderm determinant is not localized in the oocytes. Gene Expr Patterns 7:197–201
Papin C, Smith J (2000) Gradual refinement of activin-induced thresholds requires protein synthesis. Dev Biol 217:166–172
Rex M, Hilton E, Old R (2002) Multiple interactions between maternally-activated signalling pathways control Xenopus nodal-related genes. Int J Dev Biol 46(2):217–226
Rogers K, Schier A (2011) Morphogen gradients: from generation to interpretation. Ann Rev Cell Dev Biol 27(1):377–407
Saka Y, Smith J (2007) A mechanism for the sharp transition of morphogen gradient interpretation in Xenopus. BMC Dev Biol 7:47
Schohl A, Fagotto F (2002) \(\beta \)-catenin, MAPK and Smad signalling during early Xenopus development. Development 129:37–52
Schohl A, Fagotto F (2003) A role for maternal \(\beta \)-catenin in early mesoderm induction in Xenopus. EMBO J 22(13):3303–3313
Smith J, Price B, Van Nimmen K, Huylebroeck D (1990) Identification of a potent Xenopus mesoderm-inducing factor as a homologue of activin A. Nature 345(6277):729–731
Stennard F, Carnac G, Gurdon J (1996) The Xenopus T-box gene, Antipodean, encodes a vegetally localised maternal mRNA and can trigger mesoderm formation. Development 4188:4179–4188
Swiers G, Chen Y, Johnson A, Loose M (2010) A conserved mechanism for vertebrate mesoderm specification in urodele amphibians and mammals. Dev Biol 343(1–2):138–152
Takahashi S, Yokota C, Takano K, Tanegashima K, Onuma Y, Goto J, Asashima M (2000) Two novel nodal-related genes initiate early inductive events in Xenopus Nieuwkoop center. Development 127(24):5319–5329
Takahashi S, Onuma Y, Yokota C, Westmoreland J, Asashima M, Wright C (2006) Nodal-related gene Xnr5 is amplified in the Xenopus genome. Genesis 44(7):309–321
Wartlick O, Kicheva A, Gonzalez-Gaitan M (2009) Morphogen gradient formation. Cold Spring Harbor Perspect Biol 1(3):a001255
Wolpert L (1969) Positional information and the spatial pattern of cellular differentiation. J Theor Biol 25(1):1–47
Zhang J, Houston D, King M, Payne C, Wylie C, Heasman J (1998) The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell 94(4):515–524
Acknowledgments
L.E.B. was funded by the University of Nottingham Interdisciplinary Doctoral Training Centre (IDTC) in Integrative Biology. A.M.M. was supported by the Engineering and Sciences Research Council for this work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11538_2016_150_MOESM1_ESM.pdf
Supplementary Fig. S1. Numerical solutions to the Xenopus mesendoderm model, with Nodal regulation as in Xnr1/Xnr2 case, investigating the effect of VegT gradient on position of mesoderm and endoderm. Concentrations of VegT and \(\beta \)-catenin are as defined in (11) with \(a=10\) and various values of b. Parameter values were chosen as in Table 3 except \(\sigma _E=2\), colour scale used is as described in Fig. 2(c). (a) \(b=0.25\). Brachyury is expressed in the marginal zone where VegT concentrations are low and Mix is expressed at the vegetal pole where VegT is high. (b) \(b=0.75\) (i.e. VegT gradient steeper than in (a)). Note that the Mix expressing region has expanded into the marginal zone and the Brachyury expression region is smaller than in (a). (c) \(b=0.1\) (i.e. VegT gradient shallower than in (a)). Brachyury expression has widened into the vegetal region, narrowing the expression domain of Mix (pdf 1516 KB)
Appendices
Appendix 1: Model Formulation
Here we describe the formulation of our mathematical models, which are an extension of those in Middleton et al. (2009, 2013) and Brown et al. (2014). As our models include a large number of equations, we introduce the system part by part.
1.1 Nodal Signalling
We extend the Nodal signalling model developed in Middleton et al. (2013) to include regulation of Nodal by the maternal factors VegT (V) and \(\beta \)-catenin (C). The maternal factors are modelled as intracellular protein deposits, which are degraded at a constant rate, \(\mu _V\) and \(\mu _C\), respectively.
We now proceed to describe the rest of the Nodal–Antivin signalling network, first formulated by Middleton et al. (2013). Nodal and Smad families are treated as single components in our models, motivated by a single Nodal gene acting in mesendoderm formation in axolotl (Swiers et al. 2010), together with noting that the multiple Nodal genes present in Xenopus are not present in other vertebrates.
Nodal ligands (\(N^o\)) bind to free receptors (R) in a reversible reaction to become Nodal-bound receptors (\(R^\diamond \)) with \(k_N\) and \(k_{-N}\) being the association and dissociation rates of Nodal to its receptor:
Following the binding of Nodal to its receptor, intracellular Smad2 (S) becomes phosphorylated, forming PSmad2 (P). It is assumed that this occurs via the formation of an intermediate complex (\(R^{\diamond \diamond }\)). Nodal-bound receptors are rephosphorylated instantaneously after the formation of PSmad2:
PSmad2 then regulates downstream targets of the pathway, including Nodal itself, where the activation of downstream targets is modelled using a Hill function and PSmad2 is turned over at a rate \(\mu _P\).
We assume that Nodal and Antivin mRNA and extracellular proteins are degraded at rates proportional to their concentrations (\(\mu _N\),\(\mu _T\),\({\mu _N^o}\),\({\mu _T^o}\)):
Both Nodal and Antivin mRNA are assumed to be translated into protein and immediately secreted from the cell (see “Appendix 2” for further details), at a rate proportional to their concentration, with rate constants \(\delta _N\) and \(\delta _T\), respectively:
Antivin is a downstream target of Nodal signalling, which acts to antagonise Nodal signalling. This is modelled using two different mechanisms: receptor-mediated repression and heterodimer-mediated repression. In receptor-mediated repression, Antivin ligands (\(T^o\)) bind to a free receptor to form an Antivin-bound receptor (\(R^\ddagger \)) which is inactive; the rate of association and dissociation of Antivin to free receptors are \(k_T\) and \(k_{-T}\):
In heterodimer-mediated repression, an Antivin ligand binds directly to a Nodal ligand to form a Nodal–Antivin heterodimer (\(T^\ddagger \)) which is inactive, with association and dissociation rates \(l_T\) and \(l_{-T}\):
Extracellular Nodal, extracellular Antivin and Nodal–Antivin heterodimer can move between neighbouring cells with rates of transmission \(\sigma _N\), \(\sigma _T\) and \(\sigma _{T^\ddagger }\), respectively. All other species do not move between cells.
We note that the intracellular, receptor and extracellular protein concentrations are defined as the number of molecules per intracellular, cell membrane and extracellular volume, respectively. We thus need to introduce the extracellular and membrane volume fractions \(\rho =\nu _e/\nu _{j}\) and \(\nu =\nu _M/\nu _{j}\) where \(\nu _{j}\), \(\nu _E\) and \(\nu _M\) are the local intracellular, extracellular and membrane volumes, respectively.
Equations are formulated using the law of mass action, and thus, the equations governing Nodal signalling in cell j are given by:
where the term giving the production of Nodal is described in Sect. 3.1 of the main text.
1.2 FGF Signalling
The reactions involved in our formulation of the FGF signalling pathway are given in Sect. 3.2 of the main text, applying the law of mass action to (7)–(9) results in the following system of dimensional ordinary differential equations:
1.3 Xenopus Network Downstream of Nodal
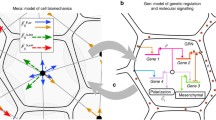
Based on the Xenopus mesendoderm shown in Fig. 1a, the time evolution of Brachyury, Goosecoid, Mix, Siamois and Lim1 in Xenopus is governed by
1.4 Axolotl Network Downstream of Nodal
Based on the axolotl mesendoderm GRN shown in Fig. 1b, the time evolution of Brachyury, Goosecoid, Mix and Lim1 in axolotl is governed by
1.5 Nondimensionalisation
We proceed by nondimensionalising (20)–(23) following the scalings used in Middleton et al. (2013) which/textbfwe summarise here. Dimensionless time (\(\tau \)) is based on the rate of turnover and secretion of intracellular Antivin, such that \(\tau \equiv \mu _N t\). The dimensionless rates of signalling (\(\hat{\sigma }_X\)), turnover (\(\hat{\mu }_Y\)) and dissociation of receptors (\(\hat{k}_{-Z}\)) are defined by \(\hat{\sigma }_X \equiv \sigma _X / \mu _N\) for \(X\in \left\{ {N,T,T^\ddagger ,E}\right\} \), \(\hat{\mu }_Y \equiv \mu _Y/ \mu _N\) for \(Y\in \left\{ {N,T,E}\right\} \) and \(\hat{k}_{-Z} \equiv k_{-Z}/ \mu _N\) for \(Z\in \left\{ {N,T,,T^\ddagger ,E,S,K}\right\} \). Dimensionless rates of secretion of Nodal, Antivin and FGF are given by \(\hat{\delta }_N \equiv \delta _N/\mu _N\) and \(\hat{\delta }_X \equiv \delta _X / \mu _N\) for \(X\in \left\{ {T,E}\right\} \). Dimensionless association rates are \(\hat{k}_s \equiv k_sS/\mu _N\), \(\hat{k}_K \equiv k_KK/\mu _N\), \(\hat{k}_Y \equiv k_YR^\bullet /\mu _N\) for \(Y\in \left\{ N,T\right\} \) and \(\hat{k}_E \equiv k_EF^\bullet /\mu _N\). The association and dissociation rates of the Nodal–Antivin complex are scaled by \(\hat{l}_T \equiv l_T R^\bullet / \left( \mu _N+\delta _N\right) \) and \(\hat{l}_{-T} \equiv l_{-T}/ \left( \mu _N+\delta _N\right) \). The nondimensional phosphorylation rate of Smad2 is \(\hat{k}_P \equiv k_P/ k_{-S}\), and the nondimensional phosphorylation rate of MAPK is \(\hat{k}_{K*}\equiv {k}_{K*} / \left( \mu _N+\delta _N\right) \). Nondimensional rates of production are: \(\hat{\lambda }_{P,N}\equiv {\lambda }_{P,N} /\mu _NR^\bullet \), \(\hat{\lambda }_{P,T}\equiv {\lambda }_{P,T} /\mu _NR^\bullet \) and \(\hat{\lambda }_{C,N}\equiv {\lambda }_{C,N} /\mu _NR^\bullet \). Dimensionless concentrations are based on the total number of receptors, such that for \(X\in \left\{ N,N^o,R^\diamond ,R^{\diamond \diamond },R\right\} \) we have \(\hat{X}=X/R^\bullet \) and for \(Y \in \left\{ E^o,K*,F,F^\diamond ,F^{\diamond \diamond }\right\} \) we have \(\hat{Y}=Y/F^\bullet \). The dimensionless concentration of P-Smad2 is \(\hat{P} \equiv k_sP / k_{-S}\), and remaining concentrations are given by \(\hat{Z}\equiv Z/ \theta _Z\), where the following are written for notational simplicity:\(\theta _G \equiv \theta _{G,B},\;\;\;\theta _B \equiv \theta _{B,E},\;\;\;\theta _E \equiv \theta _{E,B},\;\;\;\theta _L \equiv \theta _{L,G},\;\;\;\theta _M \equiv \theta _{M,B}.\) We define the following dimensionless thresholds: \(\hat{\theta }_{P,N}\equiv k_s\theta _{P,N} / k_{-S}\), \(\hat{\theta }_{P,T}\equiv \theta _{P,T} / R^\bullet \), \(\hat{\theta }_{K*,B}\equiv \theta _{K*,B} / F^\bullet \), \(\hat{\theta }_{P,B}\equiv k_S\theta _{P,B} / k_{-S}\) and \(\hat{\theta }_{P,M}\equiv k_S\theta _{P,M} / k_{-S}\). Remaining dimensionless rates of production, turnover and thresholds are defined by \(\hat{\lambda }_{Y,Z}\equiv \lambda _{Y,Z} / \theta _Z\mu _N\), \(\hat{\mu }_Z \equiv {\mu }_Z/\mu _N\) and \(\hat{\theta }_{X,Z} \equiv \theta _{X,Z} / \theta _{Z}\).
1.5.1 Parameter Sizes
We again follow the work of Middleton (2007) and Middleton et al. (2013) in making the following assumptions about the rate at which certain processes occur, based on the fact that intracellular Nodal is turned over at a faster rate than extracellular Nodal (i.e. \(\mu _N\ll \mu _{N^o}\)). It is assumed that other processes occur on a similarly fast timescale and we make the following rescalings

for \(X=\)(N,S,T,E,K), \(Y=\)(N,P,S,T,E,K), \(Z_1=\)(\(N^o\),\(T^o\),\(E^o\),\(K^*\),P), \(Z_2=(N^o,T^o,E^o)\) where \(\varepsilon =\mu _N / \mu _{N^o}\ll 1\). After applying the scalings described in this section, we defining the following for notational simplicity: \(\bar{\delta }_N = \hat{\delta }_N/\rho \), \(\bar{k}_P = \rho \hat{k}_P \hat{k}_s S^{-1}\), \(\bar{k}_{-S}=\hat{k}_{_S}(1+\hat{k}_P)-\bar{k}_P,\) \(\bar{\nu }=\nu / \rho ,\) \(\bar{\delta }_E \equiv \dfrac{\hat{\delta }_E}{\rho }\hat{\theta }_{M^*,B}\).
1.6 Dimensionless Equations
After applying the scalings described above, dropping the hats for notational simplicity, the nondimensional forms of (20) are
where \(\mathcal {F}_Y(V_j,C_j,P_j)\) is given by one of the three following functions
or
Components of the FGF signalling pathway are governed by the following nondimensional equations
The nondimensional equations governing the time evolution of Brachyury, Goosecoid, Mix, Siamois and Lim1 in Xenopus are, respectively
The nondimensional equations governing the time evolution of Brachyury, Goosecoid, Mix and Lim1 in axolotl are, respectively
Appendix 2: Modelling Transcription, Translation and Secretion of Signalling Molecules
Throughout this work, we adopt the following framework for modelling the transcription, secretion and translation of signalling molecules (other aspects, such as the association and dissociation of proteins, are modelled using the law of mass action and are described elsewhere). Let m be the concentration of some mRNA, which is transcribed at some rate \(\lambda \) and turned over at rate \(\mu _m\). Schematically, we write:
Let the intracellular and extracellular concentrations of protein which the mRNA encodes be given by p and \(p^o\). The mRNA is translated into protein at a rate \(\delta _{\mathrm{trans}}\) proportional to its concentration. mRNA is not consumed during this process. The protein can either be turned over in the cell, or secreted. Both are assumed to occur at a rate proportional to the concentration of protein in the cell, at rates given by \(\mu _{\mathrm{p}}\) and \(\delta _{\mathrm{sec}}\), respectively. We further assume that the extracellular protein is turned over at rate \(\mu _{p^o}\). Schematically, we write:
The equations governing the concentration of mRNA, intracellular protein and extracellular protein are given by:
Since this is only intended as an illustrative example, we neglect differences in volume between the extracellular and intracellular compartments (this is discussed further below). To ensure that the number of parameters in the models we develop do not become excessive, we henceforth simplify this process by assuming that the mRNA is immediately translated into protein and then secreted by the cell, so that schematically we instead write:
so that the governing equations can be written as:
We note that adopting (35) instead of (33) is equivalent (up to a scaling) to assuming that protein translation (\(\delta _{\mathrm{trans}} \)) and intracellular turnover (\(\mu _{p}\)) occur on a much faster timescale than other processes under consideration in (34). Thus, under this assumption, the level of intracellular protein p can be taken to be proportional (approximately) to the level of mRNA (i.e. \(p=\delta _{\mathrm{trans}} /\mu _p m\)), which can be directly substituted into the equation governing extracellular protein concentration in (34):
which is of the same form as 36.
Rights and permissions
About this article
Cite this article
Brown, L.E., Middleton, A.M., King, J.R. et al. Multicellular Mathematical Modelling of Mesendoderm Formation in Amphibians. Bull Math Biol 78, 436–467 (2016). https://doi.org/10.1007/s11538-016-0150-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-016-0150-8