Abstract
Multisite phosphorylation plays an important role in intracellular signaling. There has been much recent work aimed at understanding the dynamics of such systems when the phosphorylation/dephosphorylation mechanism is distributive, that is, when the binding of a substrate and an enzyme molecule results in the addition or removal of a single phosphate group and repeated binding therefore is required for multisite phosphorylation. In particular, such systems admit bistability. Here, we analyze a different class of multisite systems, in which the binding of a substrate and an enzyme molecule results in the addition or removal of phosphate groups at all phosphorylation sites, that is, we consider systems in which the mechanism is processive, rather than distributive. We show that in contrast to distributive systems, processive systems modeled with mass-action kinetics do not admit bistability and, moreover, exhibit rigid dynamics: each invariant set contains a unique equilibrium, which is a global attractor. Additionally, we obtain a monomial parametrization of the steady states. Our proofs rely on a technique of Johnston for using “translated” networks to study systems with “toric steady states,” recently given sign conditions for the injectivity of polynomial maps, and a result from monotone systems theory due to Angeli and Sontag.
Similar content being viewed by others
Notes
In sequential (de)phosphorylation, phosphate groups are added or removed in a prescribed order.
References
Anderson DF (2008) Global asymptotic stability for a class of nonlinear chemical equations. SIAM J Appl Math 68(5):1464–1476
Angeli D, De Leenheer P, Sontag ED (2007) A petri net approach to the study of persistence in chemical reaction networks. Math Biosci 210(2):598–618
Angeli D, Sontag ED (2008) Translation-invariant monotone systems, and a global convergence result for enzymatic futile cycles. Nonlinear Anal Real World Appl 9(1):128–140
Angeli D, De Leenheer P, Sontag E (2010) Graph-theoretic characterizations of monotonicity of chemical networks in reaction coordinates. J Math Biol 61(4):581–616
Aoki K, Takahashi K, Kaizu K, Matsuda M (2013) A quantitative model of ERK MAP kinase phosphorylation in crowded media. Sci Rep 3:1541. doi:10.1038/srep01541
Banaji M (2009) Monotonicity in chemical reaction systems. Dyn Syst 24(1):1–30
Banaji M, Craciun G (2009) Graph-theoretic approaches to injectivity and multiple equilibria in systems of interacting elements. Commun Math Sci 7(4):867–900
Banaji M, Mierczyński J (2013) Global convergence in systems of differential equations arising from chemical reaction networks. J Differ Equ 254(3):1359–1374
Banaji M, Pantea C (2013) Some results on injectivity and multistationarity in chemical reaction networks, preprint arXiv:1309.6771
Conradi C, Saez-Rodriguez J, Gilles E-D, Raisch J (2005) Using chemical reaction network theory to discard a kinetic mechanism hypothesis. IEEE Proc Syst Biol (now IET Syst Biol) 152(4):243–248
Conradi C, Flockerzi D, Raisch J (2008) Multistationarity in the activation of a MAPK: parametrizing the relevant region in parameter space. Math Biosci 211(1):105–131
Conradi C, Mincheva M (2014) Catalytic constants enable the emergence of bistability in dual phosphorylation. J R Soc Interface 11(95):20140158. doi:10.1098/rsif.2014.0158
Donnell P, Banaji M (2013) Local and global stability of equilibria for a class of chemical reaction networks. SIAM J Appl Dyn Syst 12(2):899–920
Donnell P, Banaji M, Marginean A, Pantea C (2014) CoNtRol: an open source framework for the analysis of chemical reaction networks. Bioinformatics 30(11):1633–1634. doi:10.1093/bioinformatics/btu063
Ellis RJ (2001) Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr Opin Struct Biol 11(1):114–119
Ellison P, Feinberg M, Ji H, Knight D (2011) Chemical reaction network toolbox. http://www.crnt.osu.edu/CRNTWin
Feinberg M (1995) Multiple steady states for chemical reaction networks of deficiency one. Arch Ration Mech Anal 132(4):371–406
Feliu E, Wiuf C (2012) Enzyme-sharing as a cause of multi-stationarity in signalling systems. J R Soc Interface 9(71):1224–1232
Feliu E, Wiuf C (2013) Simplifying biochemical models with intermediate species. J R Soc Interface 10:20130484
Flockerzi D, Holstein K, Conradi C (2014) N-site phosphorylation systems with 2N–1 steady states. Bull Math Biol 76(8):1892–1916
Gopalkrishnan M, Miller E, Shiu A (2014) A geometric approach to the global attractor conjecture. SIAM J Appl Dyn Syst 13(2):758–797
Gunawardena J (2005) Multisite protein phosphorylation makes a good threshold but can be a poor switch. PNAS 102(41):14617–14622
Gunawardena J (2007) Distributivity and processivity in multisite phosphorylation can be distinguished through steady-state invariants. Biophys J 93(11):3828–3834
Hell J, Rendall AD (2014) A proof of bistability for the dual futile cycle. Preprint. arXiv:1404.0394
Holstein K, Flockerzi D, Conradi C (2013) Multistationarity in sequential distributed multisite phosphorylation networks. Bull Math Biol 75(11):2028–2058
Horn F, Jackson R (1972) General mass action kinetics. Arch Ration Mech Anal 47(2):81–116
Johnston MD (2014) Translated chemical reaction networks. Bull Math Biol 76(6):1081–1116
Joshi B, Shiu A (2012) Simplifying the Jacobian criterion for precluding multistationarity in chemical reaction networks. SIAM J Appl Math 72(3):857–876
Manrai AK, Gunawardena J (2008) The geometry of multisite phosphorylation. Biophys J 95(12):5533–5543
Marin G, Yablonsky GS (2011) Kinetics of chemical reactions. Wiley-VCH, Wienheim
Markevich NI, Hoek JB, Kholodenko BN (2004) Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J Cell Biol 164(3):353–359
Müller S, Feliu E, Regensburger G, Conradi C, Shiu A, Dickenstein A (2013) Sign conditions for injectivity of generalized polynomial maps with applications to chemical reaction networks and real algebraic geometry. Found Comput Math (To appear)
Patwardhan P, Miller WT (2007) Processive phosphorylation: mechanism and biological importance. Cell Signal 19(11):2218–2226
Peréz Millán M, Turjanski AG (2014) MAPK’s networks and their capacity for multistationarity due to toric steady states. Preprint, arXiv:1403.6702
Peréz Millán M, Dickenstein A, Shiu A, Conradi C (2012) Chemical reaction systems with toric steady states. Bull Math Biol 74(5):1027–1065
Salazar C, Höfer T (2009) Multisite protein phosphorylation—from molecular mechanisms to kinetic models. FEBS J 276(12):3177–3198
Shinar G, Feinberg M (2012) Concordant chemical reaction networks. Math Biosci 240(2):92–113
Smith HL (1995) Monotone dynamical systems: an introduction to the theory of competitive and cooperative systems, Mathematical Surveys and Monographs, vol 41. American Mathematical Society, Providence
Stanley RP (1999) Enumerative combinatorics, vol 2, Cambridge studies in advanced mathematics, vol 62. Cambridge University Press, Cambridge
Thomson M, Gunawardena J (2009) The rational parameterisation theorem for multisite post-translational modification systems. J Theoret Biol 261(4):626–636
Thomson M, Gunawardena J (2009) Unlimited multistability in multisite phosphorylation systems. Nature 460:274–277
Wang L, Sontag ED (2008) On the number of steady states in a multiple futile cycle. J Math Biol 57(1):29–52
Acknowledgments
We thank Murad Banaji and Pete Donnell for directing us to the relevant monotone systems literature. We also thank Matthew Johnston for helpful discussions, and two conscientious referees whose comments improved this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
A. S. was supported by the NSF (DMS-1004380 and DMS-1312473). C. C. was supported in part from BMBF Grant Virtual Liver (FKZ 0315744) and the research focus dynamical systems of the state Saxony-Anhalt.
Appendix: Obtaining the Nullspace of \(A^t_\kappa \) from (17)
Appendix: Obtaining the Nullspace of \(A^t_\kappa \) from (17)
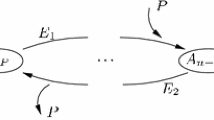
Here, we focus on the nullspace of \(A^t_\kappa \) and explain how it can be obtained by studying the directed graph underling network (17), given in Fig. 1 below.
Directed graph \(G\) underlying the translated network (17)
Notation (\(G^*\)). For a directed graph \(G\), we let \(G^*\) denote the undirected graph obtained from \(G\) by making each directed edge undirected (and allowing multiple edges in the resulting graph).
Definition 8.1
(Directed spanning tree/spanning tree rooted at node \(j\)). Let \(j\) be a node of a directed graph \(G\). A subgraph \(T\) is a spanning tree (of \(G\)) rooted at \(j\), if it satisfies the following:
-
(a)
\(T\) contains all nodes of \(G\),
-
(b)
the undirected graph \(T^*\) is acyclic and connected, and
-
(c)
for every node \(v\ne j\) of \(T\), there exists a directed path from \(v\) to \(j\).
A subgraph is a directed spanning tree of \(G\) if it is a spanning tree rooted at \(j\), for some node \(j\).
Remark 8.2
In a directed graph, a sink is a node that has no outgoing edges. For a spanning tree rooted at \(j\), the unique sink is the node \(j\). Any acyclic and connected subgraph that contains more than one sink is not a directed spanning tree.
Next, we identify the directed spanning trees of \(G\) from Fig. 1. Note that \(G\) is cyclic, and due to the unidirectional edges labeled \(k_{2n+1}\) and \(\ell _1\), \(G\) can be traversed in the clockwise direction only.
Remark 8.3
(Acyclic, connected subgraphs of \(G\) from Fig. 1). For a subgraph \(T\) of \(G\) that contains all nodes of \(G\), the undirected graph \(T^*\) is acyclic and connected if and only if \(T\) satisfies the following properties (cf. Fig. 2):
-
(i)
there is a unique node \(p\) such that \(T\) contains neither the edge \(p \rightarrow p+1\) nor the edge \(p \leftarrow p+1\) (where \(p+1:=1\) if \(p=2n+2\)).
-
(ii)
for all other nodes \(q \ne p\), exactly one of the edges \(q \rightarrow q+1\), and \(q \leftarrow q+1\) is present in \(T\).
(Color figure online) a Subgraph obtained from \(G\) by removing edges \(p \rightarrow p+1\) and \(p \leftarrow p+1\). To obtain a subgraph \(T\) for which the undirected graph \(T^*\) is acyclic and connected, choose one edge from each pair of reversible edges. By choosing all the blue edges, one obtains two directed paths ending at \(j\): one connecting the nodes \(p+1\),..., \(j-1\) to \(j\) and the other connecting \(j+1\), ..., \(n+1\) to \(j\). No choice of edges, however, will connect any of the following nodes to \(j\): \(n+2\), ..., \(2n+2\) and \(1\), ..., \(p\). Thus, any such subgraph will have at least two sinks. b Spanning tree \(T_{j,p}\) (of \(G\) from Fig. 1) rooted at \(j\); this tree consists of two paths, one from \(p\) to \(j\) (green, counterclockwise) and one from \(p+1\) to \(j\) (red, clockwise)
Now, we can determine the directed spanning trees of \(G\) (recall Definition 8.1):
Proposition 8.4
(Directed spanning trees of \(G\) from Fig. 1). For the directed graph \(G\) in Fig. 1, let \(j\) and \(p\) be integers such that
Let \(T_{j,p}\) be the subgraph of \(G\) that contains all nodes of \(G\) and for which the edges are comprised of
-
(1)
if \(j \ne n+1, 2n+2\)
-
(A)
the clockwise path from node \(p+1\) to \(j\), and
-
(B)
the counterclockwise path from \(p\) to \(j\) (cf. Fig. 2b).
-
(A)
-
(2)
if \(j=n+1\) or \(j=2n+2\), the clockwise path from node \(j+1\) to \(j\) (where \(j+1:=1\) if \(j=2n+2\)).
Then, \(T_{j,p}\) is a directed spanning tree rooted at node \(j\) that does not contain the edges \(p \rightarrow p+1\) or \(p \leftarrow p+1\) (where \(p+1:=1\) if \(p=2n+2\)). Conversely, every spanning tree of \(G\) has this form.
Proof
Assume that \(T_{j,p}\) is a subgraph as described in the proposition. By Definition 8.1 and Remark 8.3, it remains only to show that there exists a path from every node \(v\ne j\) to \(j\). Indeed, by points (1) and (4), every node belongs to a path that ends in \(j\).
Conversely, let \(T\) be a spanning tree of \(G\) rooted at \(j\). By Remark 8.3, there exists a node \(p\) such that \(T\) contains neither \(p \rightarrow p+1\) nor \( p \leftarrow p+1\), so it suffices to check that condition (45) holds and the edges of \(T\) satisfy points (1) and (4). We first assume that \(p\) violates condition (45). By symmetry between the two cases, we need only to consider the case when \(1 \le j \le n+1\) and \(p\in \{1, \dots , j-1\} \cup \{n+2, \dots , 2n+2\}\). If \(p\in \{1, \dots , j-1\}\), then there is no path in \(T\) from \(p\) to \(j\); similarly, if \(p \in \{n+2, \dots , 2n+2\}\), then there is no path from \(n+2\) to \(j\) (cf. Fig. 2). Thus, \(T\) is not a spanning tree rooted at \(j\), which is a contradiction. Thus, \(T\) must satisfy condition (45), so it remains only to show that it must satisfy points (1) and (4) as well. Indeed, in the first case (that is, if \(j \ne n+1, 2n+2\)), the paths (A) and (B) are the unique paths in \(G\) that do not use \(p \rightarrow p+1\) to reach \(j\) from \(p+1\) and \(p\), respectively, and all nodes except \(j\) lie on exactly one of these paths, so the two paths comprise the edges of \(T\). Similarly, in the remaining case (if \(j=n+1\) or \(j=2n+2\)), the clockwise path from node \(j+1\) to \(j\) is the unique path in \(G\) from \(j+1\) to \(j\), and all nodes lie along the path (note that \(j=p\) in this case). This completes the proof. \(\square \)
We note the following corollary of Proposition 8.4:
Corollary 8.5
For the directed graph \(G\) in Fig. 1, the number of spanning trees rooted at \(j\) is
-
\(n+2-j\), if \(j\in \{1, \ldots , n+1\}\)
-
\(2n+3 -j\), if \(j\in \{n+2, \ldots , 2n+2\}\).
Consequently, the number of spanning trees rooted at \(j\) is at most \(n+1\).
Now, we turn to the kernel of \(A_\kappa ^t\). In Corollary 5.1, we argued that \(\ker (A_\kappa ^t)\) is spanned by a positive vector. This is a consequence of (Thomson and Gunawardena (2009a), Lemma 2), which built on the well-known Matrix-Tree Theorem of algebraic combinatorics (Stanley 1999, §5.6) and also gives an explicit formula for this vector. For this, we need some more notation.
Notation. Following Thomson and Gunawardena (2009a), for a directed spanning tree \(T\) of an edge-labeled directed graph \(G\), we denote by \(L(T)\) the product of all edge labels in the spanning tree \(T\):
Note that \(L(T) > 0\), as it is a product of rate constants.
Proposition 8.6
Recall the spanning trees \(T_{j,p}\) of \(G\) from Fig. 1. For the matrix \(\tilde{A}_{\kappa }^t\) displayed in (20) for the translated network (17), the nullspace is spanned by the positive vector \(\rho \in {\mathbb {R}}_{+}^{2n+2}\) whose coordinates are given below
The terms \(L(T_{j,p})\) are defined in Eq. (48) below.
Proof
Proposition 8.4 and application of (Thomson and Gunawardena (2009a), Lemma 2) to \(G\) from Fig. 1.
Next, we will compute the product \(L(T_{j,p})\) associated with each spanning tree \(T_{j,p}\) of \(G\). To this end, we recall the labeling of reactions between adjacent nodes \(j\) and \(j+1\) for \(1\le j \le n-1\):

For a node \(j\) with \(n+2\le j \le 2n+1\), we write \(j\) as \(j=n+1 +i\) (so, \(1\le i \le n+1\)) and recall the labeling of reactions between adjacent nodes \(j\) and \(j+1\):

Now, we use Proposition 8.4 to compute \(L(T_{j,p})\), for a spanning tree \(T_{j,p}\) of \(G\):
-
if \(1\le j\le n\), the tree \(T_{j,p}\) splits into four paths
-
(a)
\(p+1 \rightarrow \cdots \rightarrow n+2\), with product of edge labels \(k_{2n+1}\, \prod _{i=p+1}^n k_{2i-1} = \prod _{i=p+1}^{n+1} k_{2i-1}\),
-
(b)
\(n+2\rightarrow \cdots \rightarrow 1\), with product of edge labels \(\ell _1 \prod _{i=1}^n, \ell _{2(n+1-i)+1} = \prod _{i=1}^{n+1} \ell _{2(n+1-i)+1}\),
-
(c)
\(1\rightarrow \cdots \rightarrow j\), with product of edge labels \(\prod _{i=1}^{j-1} k_{2i-1}\),
-
(d)
\(p\rightarrow \cdots \rightarrow j\), with product of edge labels \(\prod _{i=j}^{p-1} k_{2i}\).
-
(a)
-
if \(j=n+1\) (so, \(p=n+1\), by Proposition 8.4), the tree \(T_{j,p}\) splits into two paths
-
(a)
\(n+2\rightarrow \cdots \rightarrow 1\), with product of edge labels \(\prod _{i=1}^{n+1} \ell _{2(n+1-i)+1}\), as in (b) in the previous case.
-
(b)
\(1\rightarrow \cdots \rightarrow n+1\), with product of edge labels \(\prod _{i=1}^n k_{2i-1}\).
-
(a)
-
if \(n+2\le j\le 2n+1\), write \(j = n+1 + j_0\) and \(p= n+1+p_0\), and then split \(T_{j,p}\) into four paths (cf. Fig. 2b):
-
(a)
\(p+1 \rightarrow \cdots \rightarrow 1\), with product of edge labels \(\ell _1 \prod _{i=p_0+1}^n \ell _{2(n+1-i)+1} = \prod _{i=p_0+1}^{n+1} \ell _{2(n+1-i)+1}\),
-
(b)
\(1\rightarrow \cdots \rightarrow n+2\), with product of edge labels \(k_{2n+1} \prod _{i=1}^n k_{2i-1} = \prod _{i=1}^{n+1} k_{2i-1}\),
-
(c)
\(n+2 \rightarrow \cdots \rightarrow j\), with product of edge labels \(\prod _{i=1}^{j_0-1} \ell _{2(n+1-i)+1}\),
-
(d)
\(p\rightarrow \cdots \rightarrow j\), with product of edge labels \(\prod _{i=j_0}^{p_0-1} \ell _{2(n+1-i)}\).
-
(a)
-
if \(j=2n+2\) (so, \(p=2n+2\), by Proposition 8.4), the tree \(T_{j,p}\) splits into two paths
-
(a)
\(1\rightarrow \cdots \rightarrow n+2\), with product of edge labels \( \prod _{i=1}^{n+1} k_{2i-1}\), as in (b) in the previous case,
-
(b)
\(n+2\rightarrow \cdots \rightarrow 2n+2\), with product of edge labels \(\prod _{i=1}^{n}\ell _{2(n+1-i)+1}\).
-
(a)
Thus, by definition (46), we obtain for \(L(T_{j,p})\), where for \(i_1<i_0\) we adopt the standard convention \(\prod _{i=i_0}^{i_1} \alpha _i := 1\) for the empty product and, as before, \(j_0 := j-(n+1)\) and \(p_0 := p-(n+1)\):
Rights and permissions
About this article
Cite this article
Conradi, C., Shiu, A. A Global Convergence Result for Processive Multisite Phosphorylation Systems. Bull Math Biol 77, 126–155 (2015). https://doi.org/10.1007/s11538-014-0054-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-014-0054-4