Abstract
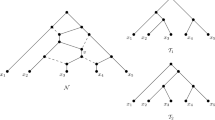
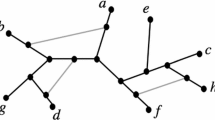
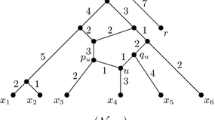
In this paper, a class of rooted acyclic directed graphs (called TOM-networks) is defined that generalizes rooted trees and allows for models including hybridization events. It is argued that the defining properties are biologically plausible. Each TOM-network has a distance defined between each pair of vertices. For a TOM-network N, suppose that the set X consisting of the leaves and the root is known, together with the distances between members of X. It is proved that N is uniquely determined from this information and can be reconstructed in polynomial time. Thus, given exact distance information on the leaves and root, the phylogenetic network can be uniquely recovered, provided that it is a TOM-network. An outgroup can be used instead of a true root.
Similar content being viewed by others
References
Bandelt, H.-J., Dress, A., 1986. Reconstructing the shape of a tree from observed dissimilarity data. Adv. Appl. Math. 7, 309–343.
Bandelt, H.-J., Dress, A., 1992. Split decomposition: A new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1, 242–252.
Baroni, M., Semple, C., Steel, M., 2004. A framework for representing reticulate evolution. Anal. Combinat. 8, 391–408.
Felsenstein, J., 1981. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Felsenstein, J., 2004. Inferring Phylogenies. Sinauer, Sunderland, MA.
Fitch, W.M., 1981. A non-sequential method for constructing trees and hierarchical classifications. J. Mol. Evol. 18, 30–37.
Dan Gusfield, Gusfield, D., 1991. Efficient algorithms for inferring evolutionary history. Networks 21, 19–28.
Gusfield, D., Eddhu, S., Langley, C., 2004a. Optimal, efficient reconstruction of phylogenetic networks with constrained recombination. J. Bioinform. Comput. Biol. 2, 173–213.
Dan Gusfield, Satish Eddhu, and Charles Langley, Gusfield, D., Eddhu, S., Langley, C., 2004b. The fine structure of galls in phylogenetic networks. INFORMS J. Comput. 16(4), 459–469.
Hasegawa, M., Kishino, H., Yano, K., 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174.
Hein, J., 1990. Reconstructing evolution of sequences subject to recombination using parsimony. Math. Biosci. 98, 185–200.
Hein, J., 1993. A heuristic method to reconstruct the history of sequences subject to recombination. J. Mol. Evol. 36, 396–405.
Huson, D.H., 1998. SplitsTree: A program for analyzing and visualizing evolutionary data. Bioinformatics 141, 68–73.
Jukes, T.H., Cantor, C.R., 1969. Evolution of protein molecules. In: Osawa, S., Honjo, T. (Eds.), Evolution of Life: Fossils, Molecules, and Culture. Springer-Verlag, Tokyo, pp. 79–95.
Kimura, M., 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120.
Li, W.-H., 1981. A simple method for construcing phylogenetic trees from distance matrices. Proc. Natl. Acad. Sci. U.S.A. 78, 1085–1089.
Vladamir Makarenkov and Pierre Legendre. Makarenkov, V., Legendre, P., 2004. From a phylogenetic tree to a reticulated network. J. Comput. Biol. 11, 195–212.
Moret, B.M.E., Nakhleh, L., Warnow, T., Linder, C.R., Tholse, A., Padolina, A., Sun, J., Timme, R., 2004. Phylogenetic networks: Modeling, reconstructibility, and accuracy. IEEE/ACM Trans. Comput. Biol. Bioinform. 1, 13–23.
Nakhleh, L., Warnow, T., Randal Linder, C., 2004. Reconstructing reticulate evolution in species—Theory and practice. In: Bourne, P.E., Gusfield, D. (Eds.), Proceedings of the Eighth Annual International Conference on Computational Molecular Biology (RECOMB '04), March 27–31, 2004, San Diego, CA. ACM, New York, pp. 337–346.
Saitou, N., Nei, M., 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sattath, S., Tversky, A., 1977. Additive similarity trees. Psychometrika 42, 319–345.
Lusheng Wang, Kaizhong Zhang, and Louxin Zhang. 2001. Wang, L., Zhang, K., Zhang, L., 2001. Perfect phylogenetic networks with recombination. J. Comput. Biol. 8, 69–78.
Willson, S.J., 2006. Unique solvability of certain hybrid networks from their distances. Ann. Combinat.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Willson, S.J. Unique Reconstruction of Tree-Like Phylogenetic Networks from Distances Between Leaves. Bull. Math. Biol. 68, 919–944 (2006). https://doi.org/10.1007/s11538-005-9044-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-005-9044-x