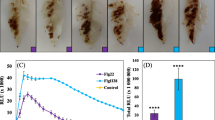
Abstract
Introduction
The rhizobacterial tomato pathogen Pseudomonas syringae pv. tomato str. DC3000 (PstDC3000), like many plant pathogenic bacteria, can elicit hypersensitive response in non-host plant cells. PstDC3000 uses a type III protein secretion system (T3SS) to deliver effector proteins.
Objectives
We compared metabolomic responses of Arabidopsis suspension cells to a wild-type PstDC3000, a T3SS deletion mutant PstDC3000D28E, and a pathogen associated molecular pattern (PAMP) flagellin’s N-terminal domain’s 22-aa peptide (flg22) to obtain metabolomics insights into the plant cell PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI).
Methods
Using targeted HPLC-MRM-MS and untargeted GC-MS approaches, we monitored qualitative and quantitative changes of 312 metabolites in central and specialized metabolic pathways in a time-course study.
Results
The overall metabolomic changes induced by the three treatments included phenylpropanoid, flavonoid, and phytohormone biosynthetic pathways, as well as primary metabolism in amino acid and sugar biosynthesis. In addition to shared metabolites, flg22, PstDC3000D28E and PstDC3000 each caused unique metabolite changes in the course of the development of PTI and ETI.
Conclusion
PstDC3000D28E triggered PTI responses were different from those of flg22. This study has not only revealed the discernible metabolomics features associated with the flg22, PstDC3000D28E and PstDC3000 treatments, but also laid a foundation toward further understanding of metabolic regulation and responses underlying plant PTI and ETI.





Similar content being viewed by others
References
Afroz, A., Zahur, M., Zeeshan, N., & Komatsu S. (2013). Plant-bacterium interactions analyzed by proteomics. Frontiers in Plant Science, 4, 21. doi:10.3389/fpls.2013.00021.
Aliferis, K. A., Faubert, D., & Jabaji, S. (2014). A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS ONE, 9, e111930.
Allwood, J. W., Clarke, A., Goodacre, R., & Mur, L. A. (2010). Dual metabolomics: A novel approach to understanding plant-pathogen interactions. Phytochemistry, 71, 590–597.
Allwood, J. W., Heald, J., Lloyd, A. J., Goodacre, R., & Mur, L. A. (2012). Separating the inseparable: The metabolomic analysis of plant-pathogen interactions. In: N. W. Hardy & R. D. Hall (Eds.), Plant metabolomics (pp. 31–49). Humana Press.
Bauer, Z., Gómez-Gómez, L., Boller, T., & Felix, G. (2001). Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. Journal of Biological Chemistry, 276, 45669–45676.
Bednarek, P., Piślewska-Bednarek, M., Svatoš, A., Schneider, B., Doubský, J., Mansurova, M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science, 323, 101–106.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate—A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 57, 289–300.
Block, A., Li, G., Fu, Z. Q., & Alfano, J. R. (2008). Phytopathogen type III effector weaponry and their plant targets. Current Opinion in Plant Biology, 11, 396–403.
Blocker, A., Komoriya, K., & Aizawa, S. I. (2003). Type III secretion systems and bacterial flagella: Insights into their function from structural similarities. Proceedings of the National Academy of Sciences of the United States of America, 100, 3027–3030.
Buell, C. R., Joardar, V., Lindeberg, M., Selengut, J., Paulsen, I. T., Gwinn, M. L., et al. (2003). The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences of the United States of America, 100, 10181–10186.
Caraux, G., & Pinloche, S. (2005). PermutMatrix: A graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics, 21, 1280–1281.
Carviel, J. L., Wilson, D. C., Isaacs, M., Carella, P., Catana, V., Golding, B., et al. (2014). Investigation of intercellular salicylic acid accumulation during compatible and incompatible Arabidopsis-Pseudomonas syringae interactions using a fast neutron-generated mutant allele of EDS5 identified by genetic mapping and whole-genome sequencing. PLoS ONE, 9, e88608.
Ceoldo, S., Toffali, K., Mantovani, S., Baldan, G., Levi, M., & Guzzo, F. (2009). Metabolomics of Daucus carota cultured cell line under stressing conditions reveals interactions between phenolic compounds. Plant Science, 176, 553–565.
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for largescale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Molecular Plant, 6, 1769–1780.
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., & Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science, 323, 95–101.
Cornelis, G. R. (2010). The type III secretion injectisome, a complex nanomachine for intracellular ‘toxin’ delivery. Biological Chemistry, 391, 745–751.
Cunnac, S., Chakravarthy, S., Kvitko, B. H., Russell, A. B., Martin, G. B., & Collmer, A. (2011). Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proceedings of the National Academy of Sciences of the United States of America, 108, 2975–2980.
ditFrey, N. F., Garcia, A. V., Bigeard, J., Zaag, R., Bueso, E., Garmier, M., et al. (2014). Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defenses. Genome Biology, 15, R87.
Duan, G., Christian, N., Schwachtje, J., Walther, D., & Ebenhöh, O. (2013). The metabolic interplay between plants and phytopathogens. Metabolites, 3, 1–23.
Dutta, B., Kanani, H., Quackenbush, J., & Klapa, M. I. (2009). Time-series integrated “omic” analyses to elucidate short-term stress-induced responses in plant liquid cultures. Biotechnology and Bioengineering, 102, 264–279.
Feng, F., & Zhou, J. M. (2012). Plant-bacterial pathogen interactions mediated by type III effectors. Current Opinion in Plant Biology, 15, 469–476.
Fiehn, O., Wohlgemuth, G., Scholz, M., Kind, T., Lee, D. Y., Lu, Y., et al. (2008). Quality control for plant metabolomics: Reporting MSI-compliant studies. Plant Journal, 53, 691–704.
Gachon, C. M., Langlois-Meurinne, M., Henry, Y., & Saindrenan, P. (2005). Transcriptional co-regulation of secondary metabolism enzymes in Arabidopsis: Functional and evolutionary implications. Plant Molecular Biology, 58, 229–245.
Galán, J. E., & Collmer, A. (1999). Type III secretion machines: Bacterial devices for protein delivery into host cells. Science, 284, 1322–1328.
Galán, J. E., & Wolf-Watz, H. (2006). Protein delivery into eukaryotic cells by type III secretion machines. Nature, 444, 567–573.
García-Alcalde, F., García-López, F., Dopazo, J., & Conesa, A. (2011). Paintomics: A web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics, 27, 137–139.
Gholami, M., Boughton, B. A., Fakhari, A. R., Ghanati, F., Mirzaei, H. H., Borojeni, L. Y., et al. (2014). Metabolomic study reveals a selective accumulation of l-arginine in the d-ornithine treated tobacco cell suspension culture. Process Biochemistry, 49, 140–147.
Grapov, D., & Newman, J. W. (2012). imDEV: A graphical user interface to R multivariate analysis tools in Microsoft Excel. Bioinformatics, 28, 2288–2290.
Guan, X., Buchholz, G., & Nick, P. (2013). The cytoskeleton is disrupted by the bacterial effector HrpZ, but not by the bacterial PAMP flg22, in tobacco BY-2 cells. Journal of Experimental Botany, 64, 1805–1816.
Hann, D. R., Domínguez-Ferreras, A., Motyka, V., Dobrev, P. I., Schornack, S., & Jehle, A. (2014). The Pseudomonas type III effector HopQ1 activates cytokinin signaling and interferes with plant innate immunity. New Phytologist, 201, 585–598.
Jones, J. D., & Dangl, J. L. (2006). The plant immune system. Nature, 444, 323–329.
Ketchum, R. E. B., Rithner, C. D., Qiu, D., Kim, Y. S., Williams, R. M., & Croteau, R. B. (2003). Taxus metabolomics: Methyl jasmonate preferentially induces production of taxoids oxygenated at C-13 in Taxus x media cell cultures. Phytochemistry, 62, 901–909.
Kim, J. K., Bamba, T., Harada, K., Fukusaki, E., & Kobayashi, A. (2007). Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. Journal of Experimental Botany, 58, 415–424.
Kim, JI., Dolan, W. L., Anderson, N. A., & Chapple, C., (2015). Indole glucosinolate biosynthesis limits phenylpropanoid accumulation in Arabidopsis thaliana. Plant Cell, 27, 1529–1546.
Kopka, J., Schauer, N., Krueger, S., Birkemeyer, C., Usadel, B., Bergmüller, E., et al. (2005). GMD@ CSB. DB: The Golm metabolome database. Bioinformatics, 21, 1635–1638.
Kruse, C., Jost, R., Lipschis, M., Kopp, B., Hartmann, M., & Hell, R. (2007). Sulfur-enhanced defence: Effects of sulfur metabolism, nitrogen supply, and pathogen lifestyle. Plant Biology, 9, 608–619.
Kuehn, H., Liberzon, A., Reich, M., Mesirov, J. P. (2008). Using GenePattern for gene expression analysis. Current protocols in bioinformatics. doi:10.1002/0471250953.bi0712s22.
Less, H., Angelovici, R., Tzin, V., & Galili, G. (2011). Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell, 23, 1264–1271.
Lindeberg, M., Cunnac, S., & Collmer, A. (2012). Pseudomonas syringae type III effector repertoires: Last words in endless arguments. Trends in Microbiology, 20, 199–208.
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L., & Fernie, A. R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nature Protocols, 1, 387–396.
Liu, D., Ford, K. L., Roessner, U., Natera, S., Cassin, A. M., Patterson, J. H., et al. (2013a). Rice suspension cultured cells are evaluated as a model system to study salt responsive networks in plants using a combined proteomic and metabolomic profiling approach. Proteomics, 13, 2046–2062.
Liu, G., Ji, Y., Bhuiyan, N. H., Pilot, G., Selvaraj, G., Zou, J., et al. (2010). Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell, 22, 3845–3863.
Liu, Y., Wang, L., Cai, G., Jiang, S., Sun, L., & Li, D. (2013b). Response of tobacco to the Pseudomonas syringae pv. tomato DC3000 is mainly dependent on salicylic acid signaling pathway. FEMS Microbiology Letters, 344, 77–85.
Liu, P., Zhang, H., Yu, B., Xiong, L., & Xia, Y. (2015). Proteomic identification of early salicylate-and flg22-responsive redox-sensitive proteins in Arabidopsis. Scientific Reports, 5, 8625. doi:10.1038/srep08625.
Mine, A., Sato, M., & Tsuda, K. (2014). Toward a systems understanding of plant-microbe interactions. Front Plant Science, 5, 423. doi:10.3389/fpls.2014.00423.
Misra, B. B., Assmann, S. M., & Chen, S. (2014). Plant single-cell and single-cell-type metabolomics. Trends Plant Science, 19, 637–646.
Misra, B. B., de Armas, E., Tong, Z., & Chen, S. (2015). Metabolomic responses of guard cells and mesophyll cells to bicarbonate. PLOS One, 10, e0144206.
Murashige, T., & Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum, 15, 473–497.
Naseem, M., Kaltdorf, M., Hussain, A., & Dandekar, T. (2013). The impact of cytokinin on jasmonate-salicylate antagonism in Arabidopsis immunity against infection with PstDC3000. Plant Signaling & Behavior, 8, e26791.
Návarová, H., Bernsdorff, F., Döring, A. C., & Zeier, J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell, 24, 5123–5141.
Noutoshi, Y., Jikumaru, Y., Kamiya, Y., & Shirasu, K. (2012). ImprimatinC1, a novel plant immune-priming compound, functions as a partial agonist of salicylic acid. Scientific Reports, 2, 705. doi:10.1038/srep00705.
Oh, C. S., & Martin, G. B. (2011). Effector-triggered immunity mediated by the pto kinase. Trends in Plant Science, 16, 132–140.
Peck, S. C., Nühse, T. S., Hess, D., Iglesias, A., Meins, F., & Boller, T. (2001). Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell, 13, 1467–1475.
Pombo, M. A., Zheng, Y., Fernandez-Pozo, N., Dunham, D. M., Fei, Z., & Martin, G. B. (2014). Transcriptomic analysis reveals tomato genes whose expression is induced specifically during effector-triggered immunity and identifies the Epk1 protein kinase which is required for the host response to three bacterial effector proteins. Genome Biology, 15, 492.
Rausch, T., & Wachter, A. (2005). Sulfur metabolism: A versatile platform for launching defence operations. Trends Plant Science, 10, 503–509.
R Development Core Team (2008). R: A language and environment for statistical computing.. Vienna, Austria: R Foundation for Statistical Computing. http://www.R-project.org.
Reich, M., Liefeld, T., Gould, J., Lerner, J., Tamayo, P., & Mesirov, J. P. (2006). GenePattern 2.0. Nature Genetics, 38, 500–501.
Rico, A., Bennett, M. H., Forcat, S., Huang, W. E., & Preston, G. M. (2010). Agroinfiltration reduces ABA levels and suppresses Pseudomonas syringae-elicited salicylic acid production in Nicotiana tabacum. PLoS ONE, 5, e8977.
Roberts, M. R. (2007). Does GABA act as a signal in plants? Hints from molecular studies. Plant Signaling & Behavior, 2, 408–409.
Robert-Seilaniantz, A., MacLean, D., Jikumaru, Y., Hill, L., Yamaguchi, S., Kamiya, Y., & Jones, J. D. (2011). The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant Journal, 67, 218–231.
Rojas, C. M., Senthil-Kumar, M., Tzin, V., & Mysore, K. S. (2014). Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Frontiers in Plant Science, 5, 17. doi:10.3389/fpls.2014.00017.
Rosli, H. G., Zheng, Y., Pombo, M. A., Zhong, S., Bombarely, A., Fei, Z., et al. (2013). Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biology, 14, R139.
Scalschi, L., Camañes, G., Llorens, E., Fernández-Crespo, E., López, M. M., García-Agustín, P., et al. (2014). Resistance inducers modulate Pseudomonas syringae pv. tomato strain DC3000 response in tomato plants. PLoS ONE, 9, e106429.
Schenk, S. T., Hernández-Reyes, C., Samans, B., Stein, E., Neumann, C., Schikora, M., et al. (2014). N-acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell, 26, 2708–2723.
Schenke, D., Boettcher, C., & Scheel, D. (2011). Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant, Cell and Environment, 34, 1849–1864.
Sokal, R. R., & Rohlf, F. J. (1995). Biometry: The principles and practice of statistics in biological research (p. 337). New York: W.H. Freeman and Company.
Soscia, C., Hachani, A., Bernadac, A., Filloux, A., & Bleves, S. (2007). Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa. Journal of Bacteriology, 189, 3124–3132.
Stein, S. E. (1999). An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. Journal of the American Society for Mass Spectrometry, 10, 770–781.
Sumner, L. W., Lei, Z., Nikolau, B. J., & Saito, K. (2014). Modern plant metabolomics: Advanced natural product gene discoveries, improved technologies, and future prospects. Natural Product Reports, 32, 212–229.
Thilmony, R., Underwood, W., & He, S. Y. (2006). Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157: H7. Plant Journal, 46, 34–53.
Trdá, L., Fernandez, O., Boutrot, F., Héloir, M. C., Kelloniemi, J., Daire, X., et al. (2014). The grapevine flagellin receptor VvFLS2 differentially recognizes flagellin-derived epitopes from the endophytic growth-promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytologist, 201, 1371–1384.
Tsuda, K., Mine, A., Bethke, G., Igarashi, D., Botanga, C. J., Tsuda, Y., et al. (2013). Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genetics, 9, e1004015.
Tsutsui, T., Nakano, A., & Ueda, T. (2015). The plant-specific RAB5 GTPase ARA6 is required for starch and sugar homeostasis in Arabidopsis thaliana. Plant and Cell Physiology, 56, 1073–1083.
Vargas, P., Farias, G. A., Nogales, J., Prada, H., Carvajal, V., Barón, M., et al. (2013). Plant flavonoids target Pseudomonas syringae pv. tomato DC3000 flagella and type III secretion system. Environmental Microbiology Reports, 5, 841–850.
Villela-Dias, C., Camillo, L. R., de Oliveira, G. A., Sena, J. A., Santiago, A. S., de Sousa, S. T., et al. (2014). Nep1-like protein from Moniliophthora perniciosa induces a rapid proteome and metabolome reprogramming in cells of Nicotiana benthamiana. Physiologia Plantarum, 150, 1–17.
Walley, J. W., Kliebenstein, D. J., Bostock, R. M., & Dehesh, K. (2013). Fatty acids and early detection of pathogens. Current Opinion in Plant Biology, 16, 520–526.
Xia, J. G., Psychogios, N., Young, N., & Wishart, D. S. (2009). MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Research, 37, W652–W660.
Zeier, J. (2013). New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant, Cell and Environment, 36, 2085–2103.
Zhou, J. M., & Chai, J. (2008). Plant pathogenic bacterial type III effectors subdue host responses. Current Opinion in Microbiology, 11, 179–185.
Zipfel, C., & Robatzek, S. (2010). Pathogen-associated molecular pattern-triggered immunity: veni, vidi…? Plant Physiology, 154, 551–554.
Acknowledgments
This work was supported by the U.S. National Science Foundation grant NSF-MCB-1158000 to SC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no potential conflicts of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Misra, B.B., de Armas, E. & Chen, S. Differential metabolomic responses of PAMP-triggered immunity and effector-triggered immunity in Arabidopsis suspension cells. Metabolomics 12, 61 (2016). https://doi.org/10.1007/s11306-016-0984-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-016-0984-y