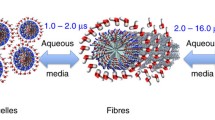
Abstract
The influence of amino acid sequence on the secondary structure of peptide amphiphile (PAs) cylindrical micelles and fibers that are self-assembled in solution is studied using molecular dynamics simulations. Simulations for two choices of PAs were performed, starting with structures that have the correct overall shape, but which restructure considerably during the simulation, with one fiber being composed of valine rich PAs and the other of alanine rich PAs. Self-assembly is similar in both simulations, with stable fibers (diameter is 7.7–8 nm) obtained after 40 ns. We find that the valine rich PA fiber has a higher β-sheet population than the alanine rich fiber, and that the number of hydrogen bonds is higher. This behavior of the valine rich fiber is consistent with experimental measurements of higher stiffness, and it shows that stiffness can be varied while still maintaining self-assembly.





Similar content being viewed by others
References
Andersen HC (1983) Rattle—a velocity version of the shake algorithm for molecular-dynamics calculations. J Comput Phys 52:24
Behanna HA, Donners J, Gordon AC, Stupp SI (2005) Coassembly of amphiphiles with opposite peptide polarities into nanofibers. J Am Chem Soc 127:1193
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an n.Log(n) method for Ewald sums in large systems. J Chem Phys 98:10089
Feller SE, Zhang YH, Pastor RW, Brooks BR (1995) Constant-pressure molecular-dynamics simulation: the Langevin piston method. J Chem Phys 103:4613
Frishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Proteins 23:566
Grubmuller H (1996) Solvate. Theoretical Biophysics Group, Institute for Medical Optics, Ludwig-Maximilian University, Munich
Hartgerink JD, Beniash E, Stupp SI (2001) Self-assembly and mineralization of peptide-amphiphile nanofibers. Science 294:1684
Hartgerink JD, Beniash E, Stupp SI (2002) Peptide-amphiphile nanofibers: a versatile scaffold for the preparation of self-assembling materials. Proc Natl Acad Sci USA 99:5133
Heinig M, Frishman D (2004) STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res 32:W500
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38
IUPAC (1997) Compendium of chemical terminology, 2nd edn. Blackwell Scientific Publications, Oxford
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926
Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K (1999) NAMD2: greater scalability for parallel molecular dynamics. J Comput Phys 151:283
Kim CWA, Berg JM (1993) Thermodynamic beta-sheet propensities measured using a zinc-finger host peptide. Nature 362:267
Langer R, Vacanti JP (1993) Tissue engineering. Science 260:920
Lee O-S, Stupp SI, Schatz GC (2011) Atomistic molecular dynamics simulations of peptide amphiphile self-assembly into cylindrical nanofibers. J Am Chem Soc 133:3677
Levitt M (1978) Conformational preferences of amino-acids in globular proteins. Biochemistry 17:4277
Mackerell AD, Wiorkewiczkuczera J, Karplus M (1995) An all-atom empirical energy function for the simulation of nucleic-acids. J Am Chem Soc 117:11946
Manning MC, Illangasekare M, Woody RW (1988) Circular-dichroism studies of distorted alpha-helices, twisted beta-sheets, and beta-turns. Biophys Chem 31:77–86
Martyna GJ, Tobias DJ, Klein ML (1994) Constant-pressure molecular-dynamics algorithms. J Chem Phys 101:4177
Meyers M, Chawla K (2009) Mechanical behavior of materials. Cambridge University Press, Cambridge
Paramonov SE, Jun HW, Hartgerink JD (2006a) Self-assembly of peptide-amphiphile nanofibers: the roles of hydrogen bonding and amphiphilic packing. J Am Chem Soc 128:7291
Paramonov SE, Jun HW, Hartgerink JD (2006b) Modulation of peptide-amphiphile nanofibers via phospholipid inclusions. Biomacromolecules 7(1):24–26
Pashuck ET, Cui HG, Stupp SI (2010) Tuning supramolecular rigidity of peptide fibers through molecular structure. J Am Chem Soc 132:6041
Salemme FR (1983) Structural-properties of protein beta-sheets. Prog Biophys Mol Biol 42:95
Stupp SI, Donners J, Li LS, Mata A (2005) Expanding frontiers in biomaterials. MRS Bull 30:864
Tashiro K, Tadokoro H (1981) Calculation of 3-dimensional elastic-constants of polymer crystals. 3. Alpha-forms and gamma-forms of nylon-6. Macromolecules 14:781
Tashiro K, Kobayashi M, Tadokoro H (1978a) Calculation of 3-dimensional elastic-constants of polyer crystals. 1. Method of calculation. Macromolecules 11:908
Tashiro K, Kobayashi M, Tadokoro H (1978b) Calculation of 3-dimensional elastic-constants of polymer crystals. 2. Application to orthorhombic polyethylene and polyvynyl-alcohol. Macromolecules 11:914
Tysseling VM, Sahni V, Pashuck ET, Birch D, Hebert A, Czeisler C, Stupp SI, Kessler JA (2010) Self-assembling peptide amphiphile promotes plasticity of serotonergic fibers following spinal cord injury. J Neurosci Res 88:3161
Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, Kessler JA (2008) Self-assembling nanofibers inhibit glial scar formation and promote axon elongation after spinal cord injury. J Neurosci 28:3814
Zhao Y, Yokoi H, Tanaka M, Kinoshita T, Tan TW (2008) Self-assembled pH-responsive hydrogels composed of the RATEA16 peptide. Biomacromolecules 9:1511
Acknowledgments
This research was supported by the National Science Foundation (grant CHE-1147335), and by the DOE NERC EFRC (DE-SC0000989).
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the topical collection on nanomaterials in energy, health and environment
Rights and permissions
About this article
Cite this article
Lee, OS., Liu, Y. & Schatz, G.C. Molecular dynamics simulation of β-sheet formation in self-assembled peptide amphiphile fibers. J Nanopart Res 14, 936 (2012). https://doi.org/10.1007/s11051-012-0936-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11051-012-0936-z