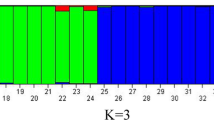
Abstract
Molecular genetic polymorphism in three species and four subspecies of crested wheatgrass, Agropyron, was studied using 56K diversity array technology (DArT), and the results confirmed with four selected SNP Amplifluor markers. In total, 82 accessions from three species—A. desertorum, A. fragile, and two subspecies of A. cristatum (ssp. cristatum and ssp. pectinatum)—were collected from various regions of Kazakhstan or ordered from Genebank in Russia, for morphological taxonomy and molecular phylogenetic analyses. In the DArT clone analysis, two Agropyron species with narrow linear spikes, A. fragile and A. desertorum, were found to be genetically similar and fell within a single clade (A). Both species share similar eco-geographical origins. All samples of A. cristatum including the two subspecies, ssp. pectinatum and ssp. cristatum, which have short broad spikes, were interspersed within two other clades, B and C, more genetically distanced from the other species. Four SNP Amplifluor markers developed for genetic fragments on different chromosomes confirmed the distinction between the studied species. These results, derived from multiple molecular markers, suggest that the morphological taxonomy of these Agropyron species should be re-considered carefully in the future.



Similar content being viewed by others
References
Bogdan VS (1937) Crested wheatgrass. Proceed Krasnodar Breeding Station.137-150. (In Russian).
Bukhteeva AV, Malyshev LL, Dzubekno NI, Kochegina AA (2016) Genetic resources of сrested wheatgrass—Agropyron Gaerth. VIR, St-Petersburg, 268 p (In Russian)
Che YH, Yang YP, Yang XM, Li XQ, Li LH (2011) Genetic diversity between ex situ and in situ samples of Agropyron cristatum (L.) Gaertn. based on simple sequence repeat molecular markers. Crop Past Sci 62(8):639–644. https://doi.org/10.1071/CP11065
Che Y, Yang Y, Yang X, Li X, Li L (2015) Phylogenetic relationship and diversity among Agropyron Gaertn. germplasm using SSRs markers. Plant Syst Evol 301:163–170. https://doi.org/10.1007/s00606-014-1062-4
Chen SY, Ma X, Zhang XQ, Huang LK, Zhou JN (2013) Genetic diversity and relationships among accessions of five crested wheatgrass species (Poaceae: Agropyron) based on gliadin analysis. Genet Mol Res 12(4):5704–5713. https://doi.org/10.4238/2013.November.18.19
Cherukuri DP, Gupta SK, Charpe A, Koul S, Prabhu KV, Singh RB, Haq QMR, Chauhan SVS (2003) Identification of a molecular marker linked to an Agropyron elongatum-derived gene Lr19 for leaf rust resistance in wheat. Plant Breed 122(3):204–208
Gascuel O (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14(7):685–695. https://doi.org/10.1093/oxfordjournals.molbev.a025808
Giancola S, McKhann HI, Bérard A, Camilleri C, Durand S, Libeau P et al (2006) Utilization of the three high-throughput SNP genotyping methods, the GOOD assay, Amplifluor and TaqMan, in diploid and polyploid plants. Theor Appl Genet 112(6):1115–1124. https://doi.org/10.1007/s00122-006-0213-6
Howard EL, Whittock SP, Jakše J, Carling J, Matthews PD, Probasco G, Henning JA, Darby P, Cerenak A, Javornik B, Kilian A, Koutoulis A (2011) High-throughput genotyping of hop (Humulus lupulus L.) utilising diversity arrays technology (DArT). Theor Appl Genet 122:1265–1280. https://doi.org/10.1007/s00122-011-1529-4
Hu ZM, Wang RRC, Larson SR, Palazzo AJ, Asay KH, Chatterton NJ (2001) Selection response for molecular markers associated with anthocyanin coloration and low-temperature growth traits in crested wheatgrasses. Canad J Plant Sci 81(4):665–671
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23(2):254–267. https://doi.org/10.1093/molbev/msj030
James KE, Schneider H, Ansell SW, Evers M, Robba L, Uszynski G, Pedersen N, Newton AE, Russell SJ, Vogel JC, Kilian A (2008) Diversity arrays technology (DArT) for pan-genomic evolutionary studies of non-model organisms. PLoS One 3(2):e1682. https://doi.org/10.1371/journal.pone.0001682
Khripin Y (2006) High-throughput genotyping with energy transfer-labeled primers. In: Didenko VV (ed) Methods in molecular biology, Vol 335, Fluorescent energy transfer nucleic acid probes: designs and protocols. Humana Press Inc, Totowa, NJ, pp 215–240
Kilian A, Huttner E, Wenzl P, Jaccoud D, Carling J et al (2005) The fast and the cheap: SNP and DArT-based whole genome profiling for crop improvement. In: Tuberosa R, Phillips RL, Gale M (eds) In the wake of the double helix: from the green revolution to the gene revolution, Proceedings of the International Congress. Avenue media, Bologna, pp 443–461
Kroc M, Koczyk G, Święcicki W, Kilian A, Nelson MN (2014) New evidence of ancestral polyploidy in the Genistoid legume Lupinus angustifolius L. (narrow-leafed lupin). Theor Appl Genet 127:1237–1249. https://doi.org/10.1007/s00122-014-2294-y
Kruppa K, Türkösi E, Mayer M, Tóth V, Vida G, Szakács É, Molnár-Láng M (2016) McGISH identification and phenotypic description of leaf rust and yellow rust resistant partial amphiploids originating from a wheat × Thinopyrum synthetic hybrid cross. J Appl Genet 57:427–437. https://doi.org/10.1007/s13353-016-0343-8
Lu XP, Xiao BG, Li YP, Gui YJ, Wang Y, Fan LJ (2013) Diversity arrays technology (DArT) for studying the genetic polymorphism of flue-cured tobacco (Nicotiana tabacum). J Zhejiang Univ Sci B (Biomed and Biotechnol) 14(7):570–577. https://doi.org/10.1631/jzus.B1200227
Lu Y, Yao M, Zhang J, Song L, Liu W, Yang X, Li X, Li L (2016) Genetic analysis of a novel broad-spectrum powdery mildew resistance gene from the wheat-Agropyron cristatum introgression line Pubing 74. Planta 244(3):713–723. https://doi.org/10.1007/s00425-016-2538-y
Majidi MM, Mirlohi A (2010) Genetic similarities among Iranian populations of Festuca, Lolium, Bromus and Agropyron using amplified fragments length polymorphism (AFLP) markers. Iran J Biotechnol 8(1): 16–23
Mellish A, Coulman B, Ferdinandez Y (2002) Genetic relationships among selected crested wheatgrass cultivars and species determined on the basis of AFLP markers. Crop Sci 42(5):1662–1668
Ochoa V, Madrid E, Said M, Rubiales D, Cabrera A (2015) Molecular and cytogenetic characterization of a common wheat-Agropyron cristatum chromosome translocation conferring resistance to leaf rust. Euphytica 201(1):89–95. https://doi.org/10.1007/s10681-014-1190-5
Olukolu BA, Mayes S, Stadler F, Ng NQ, Fawole I, Dominique D, Azam-Ali SN, Abbott AG, Kole C (2012) Genetic diversity in Bambara groundnut (Vigna subterranean (L.) Verdc.) as revealed by phenotypic descriptors and DArT marker analysis. Genet Resour Crop Evol 59:347–358. https://doi.org/10.1007/s10722-011-9686-5
Przyborowski JA, Sulima P, Kuszewska A, Załuski D, Kilian A (2013) Phylogenetic relationships between four Salix L. species based on DArT markers. Int J Mol Sci 14:24113–24125. https://doi.org/10.3390/ijms141224113
Raman H, Raman R, Nelson MN, Aslam MN, Rajasekaran R, Wratten N, Cowling WA, Kilian A, Sharpe AG, Schondelmaier J (2012) Diversity array technology markers: genetic diversity analyses and linkage map construction in rapeseed (Brassica napus L.) DNA Res 19:51–65. https://doi.org/10.1093/dnares/dsr041
Rickert AM, Borodina TA, Kuhn EJ, Lehrach H, Sperling S (2004) Refinement of single-nucleotide polymorphism genotyping methods on human genomic DNA: amplifluor allele-specific polymerase chain reaction versus ligation detection reaction-TaqMan. Anal Biochem 330:288–297. https://doi.org/10.1016/j.ab.2004.03.035
Roorkiwal M, von Wettberg EJ, Upadhyaya HD, Warschefsky E, Rathore A, Varshney RK (2014) Exploring germplasm diversity to understand the domestication process in Cicer spp. using SNP and DArT markers. PLoS One 9(7):e102016. https://doi.org/10.1371/journal.pone.0102016
Rutherford S, Wilson PG, Rossetto M, Bonser SP (2015) Phylogenomics of the green ash eucalypts (Myrtaceae): a tale of reticulate evolution and misidentification. Aust Syst Bot 28:326–354. https://doi.org/10.1071/SB15038
Schachermayr GM, Messmer MM, Feuillet C, Winzeler H, Winzeler M, Keller B (1995) Identification of molecular markers linked to the Agropyron elongatum-derived leaf rust resistance gene Lr24 in wheat. Theor Appl Genet 90(7–8):982–990
Shanjani PS, Jafari AA, Calagari M (2013) Genetic variation among wild and cultivated Agropyron desertorum populations based on total protein profiles and phenotypic traits. New Zeal J Crop Hort Sci 41(3):117–134. https://doi.org/10.1080/01140671.2013.793203
Sharp PJ, Kreis M, Shewry PR, Gale MD (1988) Location of β-amylase sequences in wheat and its relatives. Theor Appl Genet 75:286–290
Shavrukov Y, Zhumalin A, Serikbay D, Botayeva M, Otemisova A, Absattarova A, Sereda G, Sereda S, Shvidchenko V, Turbekova A, Jatayev S, Lopato S, Soole K, Langridge P (2016) Expression level of the DREB2-type gene, identified with Amplifluor SNP markers, correlates with performance and tolerance to dehydration in bread wheat cultivars from Northern Kazakhstan. Front Plant Sci 7:1736. https://doi.org/10.3389/fpls.2016.01736
Thudi M, Upadhyaya HD, Rathore A, Gaur PM, Krishnamurthy L, Roorkiwal M, Nayak SN, Chaturvedi SK, Basu PS, Gangarao NVPR, Fikre A, Kimurto P, Sharma PC, Sheshashayee MS, Tobita S, Kashiwagi J, Ito O, Killian A, Varshney RK (2014) Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS One 9(5):e96758. https://doi.org/10.1371/journal.pone.0096758
Yang S, Pang W, Ash G, Harper J, Carling J, Wenzl P, Huttner E, Zong X, Kilian A (2006) Low level of genetic diversity in cultivated Pigeonpea compared to its wild relatives is revealed by diversity arrays technology. Theor Appl Genet 113:585–595. https://doi.org/10.1007/s00122-006-0317-z
Ye X, Lu Y, Liu W, Chen G, Han H, Zhang J, Yang X, Li X, Gao A, Li L (2015) The effects of chromosome 6P on fertile tiller number of wheat as revealed in wheat-Agropyron cristatum chromosome 5A/6P translocation lines. Theor Appl Genet 128(5):797–811. https://doi.org/10.1007/s00122-015-2466-4
Yilmaz R, Cabi E, Dogan M (2014) Molecular analyses of the genera Eremopyrum (Ledeb.) Jaub. & Spach and Agropyron Gaertner (Poaceae) by PCR methods. Pak J Bot 46(3):769–774
Yu X, Li X, Ma Y, Yu Z, Li Z (2012) A genetic linkage map of crested wheatgrass based on AFLP and RAPD markers. Genome 55(4):327–335. https://doi.org/10.1139/G2012-014
Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A (2004) A diversity arrays technology (DArT) for whole-genome profiling of barley. PNAS 101(26):9915–9920. https://doi.org/10.1073/pnas.0401076101
White J, Law JR, MacKay I, Chalmers KJ,·Smith JSC, Kilian A, Powell W (2008) The genetic diversity of UK, US and Australian cultivars of Triticum aestivum measured by DArT markers and considered by genome. Theor Appl Genet 116:439–453. https://doi.org/10.1007/s00122-007-0681-3.
Zhang LY, Marchand S, Tinker NA, Belzile F (2009) Population structure and linkage disequilibrium in barley assessed by DArT markers. Theor Appl Genet 119:43–52. https://doi.org/10.1007/s00122-009-1015-4
Zhang Y, Zhang J, Huang L, Gao A, Zhang J, Yang X (2015a) A high-density genetic map for P genome of Agropyron Gaertn. based on specific-locus amplified fragment sequencing (SLAF-seq). Planta 242(6):1335–1347. https://doi.org/10.1007/s00425-015-2372-7
Zhang J, Zhang J, Liu W, Han H, Lu Y, Yang X, Li X, Li L (2015b) Introgression of Agropyron cristatum 6P chromosome segment into common wheat for enhanced thousand-grain weight and spike length. Theor Appl Genet 128(9):1827–1837. https://doi.org/10.1007/s00425-015-2372-7
Zhang J, Liu W, Han H, Song L, Bai L, Gao Z, Zhang Y, Yang X, Li X, Gao A, Li L (2015c) De novo transcriptome sequencing of Agropyron cristatum to identify available gene resources for the enhancement of wheat. Genomics 106(2):129–136. https://doi.org/10.1016/j.ygeno.2015.04.003
Zhang J, Zhang J, Liu W, Wu X, Yang X, Li X, Lu Y, Li L (2016) An intercalary translocation from Agropyron cristatum 6P chromosome into common wheat confers enhanced kernel number per spike. Planta 244(4):853–864. https://doi.org/10.1007/s00425-015-2372-7
Zhou S, Yan B, Li F, Zhang J, Zhang J, Ma H, Liu W, Lu Y, Yang X, Li X, Liu X, Li L (2017a) RNA-seq analysis provides the first insights into the phylogenetic relationship and interspecific variation between Agropyron cristatum and wheat. Front Plant Sci 8:1644. https://doi.org/10.3389/fpls.2017.01644
Zhou S, Zhang J, Che Y, Liu W, Lu Y, Yang X, Li X, Jia J, Liu X, Li L (2017b) Construction of Agropyron Gaertn. genetic linkage maps using a wheat 660K SNP array reveals a homoeologous relationship with the wheat genome. Plant Biotechnol J. https://doi.org/10.1111/pbi.12831
Acknowledgements
We wish to thank the staff and students of S. Seifullin Kazakh AgroTechnical University, Astana (Kazakhstan) for their support in this research. We also thank Carly Schramm for critical comments in the manuscript.
Funding
Research project no. 3733/GF4 has been supported by Ministry of Education and Science (Kazakhstan).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 23 kb)
Rights and permissions
About this article
Cite this article
Absattar, T., Absattarova, A., Fillipova, N. et al. Diversity array technology (DArT) 56K analysis, confirmed by SNP markers, distinguishes one сrested wheatgrass Agropyron species from two others found in Kazakhstan. Mol Breeding 38, 37 (2018). https://doi.org/10.1007/s11032-018-0792-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0792-3