Abstract
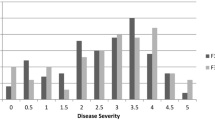
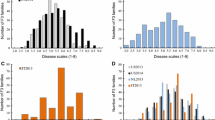
A pepper (Capsicum annuum) inbred line ‘BJ0747-1-3-1-1’ was found to exhibit resistance to a cucumber mosaic virus (CMV) local isolate (CMV-HB). In order to exploit the genetic mechanism of CMV resistance in this pepper line, inheritance and genetic linkages of CMV resistance were studied. The CMV-resistant ‘BJ0747-1-3-1-1’ (P1) was crossed to the susceptible line ‘XJ0630-2-1-2-1-1’ (P2), and the F2 and back-cross populations (B1 and B2) were generated for segregation analysis following a mixed inheritance model. Analysis of the segregation data revealed that CMV resistance in ‘BJ0747-1-3-1-1’ is controlled by two partially additive-dominant major genes and additive-dominant polygenes. Analysis of the results from two growth seasons also identified two stable and major QTLs for CMV resistance that collectively explained 55 % of the trait variation. One of the two major QTLs was found on linkage group 8 (LG8) between markers TS52 and HpmsE1-43, and accounted for 37.7–43.5 % of the variation in the two inoculation experiments. The other QTL on LG4 between markers UBC843 and S74 was found to be responsible for 10.7–11.2 % of the trait variation. QTLs with minor effects on CMV resistance were also identified. This work provides an example for genetic analysis and QTL mapping of other disease-resistance genes in pepper and the findings should be useful for molecular marker-assisted breeding programs that aim at developing disease-resistant cultivars in peppers.


Similar content being viewed by others
References
Akaike H (1977) On the entropy maximum principle. In: Krishnaiah PR (ed) Applications of statistics. North-Holland Publishing Company, Amsterdam, pp 27–41
Caranta C, Palloix A, Lefebvre V, Daubeze AM (1997) QTLs for a component of partial resistance to cucumber mosaic virus in pepper: restriction of virus installation in host-cells. Theor Appl Genet 94:431–438
Caranta C, Pxiege S, Lefebvre V, Daubeze AM, Thabuis A, Palloix A (2002) QTLs involved in the restriction of Cucumber mosaic virus (CMV) long-distance movement in pepper. Theor Appl Genet 104:586–591
Chaim BA, Grube RC, Lapidot M, Jahn M, Paran I (2001) Identification of quantitative trait loci associated with resistance to Cucumber mosaic virus in Capsicum annuum. Theor Appl Genet 102:1213–1220
Chen H, Wang S, Xing Y, Xu C, Hayes PM, Zhang Q (2003) Comparative analyses of genomic locations and race specificities of loci for quantitative resistance to Pyricularia grisea in rice and barley. Proc Natl Acad Sci USA 100(5):2544–2549
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Dong N, Li C, Wang Q, Ai N, Hu G, Zhang J (2010) Mixed inheritance of earliness and its related traits of short-season cotton under different ecolodical environments. Cotton Sci 22(4):304–311 (in Chinese)
Elston RC (1984) The genetic analysis of quantitative trait differences between two homozygous lines. Genetics 108:733–744
Fu S, Qi C (2009) Major gene plus polygene inheritance of oil contention Brassica napus L. Jiangsu J Agric Sci. 25(4):731–736 (in Chinese)
Gai J, Wang J (1998) Identification and estimation of a QTL model and its effects. Theor Appl Genet 97:1162–1168
Gai J, Zhang Y, Wang J (2003) Genetic system of quantitative traits in plants. Science Press, Beijing, pp 230–236 (in Chinese)
Grube RC, Zhang Y, Murphy JF, Loaiza-Figueroa F, Lackney VK, Provvidenti R, Jahn M (2000) New source of resistance to Cucumber mosaic virus in Capsicum frutescens. Plant Dis 84:885–891
Gu H, Qi C (2008) Genetic analysis of lodging resistance with mixed model of major gene plus polygene in Brassica napus L. Acta Agron Sinica 34(3):376–381
Huang S, Zhang B, Dan M, Cardle Linda, Yang G, Guo J (2000) Development of pepper SSR markers from sequence databases. Euphytica 117:163–167
Kang W, Hoang N, Yang H, Kwon J, Jo S, Seo J, Kim K, Choi D, Kang B (2010) Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L.). Theor Appl Genet 120:1587–1596
Kong F (2006) Quantative genetics in plants. China Agricultural University Press, Beijing, China
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Lapidot M, Paran I, Ben-Joseph R, Ben-Harush S, Pilowsky M, Cohen S, Shifris C (1997) Tolerance to Cucumber mosaic virus in pepper: development of advanced breeding lines and evaluation of virus level. Plant Dis 81:185–188
Lee JM, Nahm SH, Kim YM, Kim BD (2004) Characterization and molecular genetic mapping of microsatellite loci in pepper. Theor Appl Genet 108:619–627
Levine DM, Ramsey PP, Smidt RK (2001) Applied statistics for engineers and scientists: Using Microsoft Excel and MINITABLE. Prentice-Hall, Upper Saddle River, NJ
Li Y, Zhu Z, Zhang Y, Zhao L, Wang C (2008) Genetic analysis of rice false smut resistance using major gene plus polygene mixed genetic model. Acta Agron Sinica 34(10):1728–1733
Lincoln S, Daly M, Lander E (1992) Constructing genetic maps with mapmaker/EXP3.0. Whitehead Institute Techn. Rep, 3rd edn. Whitehead Institute, Cambridge
Luo X, Si L, Yin W (2009) Inheritance on major gene plus poly·genes of yellow line and Fruit length ratio of cucumber. Acta Agric Boreali Sinica 23(2):88–91
Minamiyama Y, Tsuro M, Hirai M (2006) An SSR-based linkage map of Capsicum annuum. Mol Breed 18:157–169
Mimura Y, Inoue T, Minamiyama Y, Kubo N (2012) An SSR-based genetic map of pepper (Capsicum annuum L.) serves as an anchor for the alignment of major pepper maps. Breed Sci 62:93–98
Murray MG, Thompson FW (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–4326
Nagy I, Stágel A, Sasvári Z, Röder M, Ganal M (2007) Development, characterization, and transferability to other Solanaceae of microsatellite markers in pepper (Capsicum annuum L.). Genome 50(7):668–688
Palukaitis P, Roossinck MJ, Dietzgen RG, Francki RIB (1992) Cucumber mosaic virus. Adv Virus Res 41:281–341
Pochard E, Daubeze AM (1989) Progressive construction of a polygenic resistance to Cucumber mosaic virus in the pepper. In: Proceedings of Eucarpia meeting on genetics and breeding of Capsicum and Eggplant, 7th, Kragujevac, Yugoslavia, pp 189–192
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Singh J, Thakur MR (1977) Genetics of resistance to tobacco mosaic virus, cucumber mosaic virus and leaf curl virus in hot pepper (Capsicum annuum L.). In: 3rd Eucarpia Meeting, Montfavet, France, pp 119–123
Suzuki K, Kuroda T, Miura Y, Muria J (2003) Screening and weld traits of virus resistant source in Capsicum spp. Plant Dis 87:779–783
Thomas WTB, Powell W, Waugh R, Chalmers KJ, Barua UM, Jack P, Lea V, Forster BP, Swanston JS, Ellis RP, Hanson PR, Lance RCM (1995) Detection of quantitative trait loci for agronomic, yield, grain and disease characters in spring barley (Hordeum vulgare L.). Theor Appl Genet 91(6–7):1037–1047
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Wang J (1996) Studies on identification of major-polygene mixed inheritance of quantitative traits and estimation of genetic parameters. Doctorate dissertation, Department of Plant Breeding and Biometrics, Nanjing Agricultural University (in Chinese)
Wang J, Gai J (1997) Identification of major gene and polygene mixed inheritance and estimation of genetic parameters in F2 progeny. Chin J Genet 24:181–190
Wang J, Gai J (2001) Mixed inheritance model for resistance to agromyzid beanfly (Melanagromyza sojae Zehntner) in soybean. Euphytica 122:9–18
Wang J, Wang J, Zhu L, Gai J (2000) Major-polygene effect analysis of resistance to bacterial blight (Xanthomonas campestris pv.orzae) in rice. Acta Genet Sinica 27(1):34–38
Wang S, Basten CJ, Gaffney P, Zeng ZB (2004) Windows QTL cartographer version 2.0. Statistical genetics. North Carolina State University, USA
Yan S, Ju J, Xu X, Cui Y (1996) Genetic analysis on resistance of sweet (hot) pepper to CMV. Acta Hortic Sinica 23(1):45–48 (In Chinese)
Yan L, Luo L, Feng Z, Li X, Lou Q, Chen J (2009) Analysis on mixed maj or gene and polygene inheritance of parthenocarpy in monoecious cucumber (Cucumis sativus L). Acta Botanica Boreali Occidentalia Sinica 29(6):1122–1126 (in Chinese)
Yao M, Wang F, Ye Z (2009) Subgroup identification and strain analysis of the cucumber mosaic virus infecting pepper plant in Hubei Province. J Huazhong Agric Univ 28(4):472–475 (In Chinese)
Yu X, Kong Q, Wang Y (2000) Inheritance of resistance to cucumber mosaic virus pepper isolate (CMV-P) in Capsicum annum L. J Shenyang Agric Univ 31(2):169–171 (In Chinese)
Zhang Y, Gai J (2000) Identification of two major genes plus polygenes mixed inheritance model of quantitative trait in B1 and B2, and F2. J Biomath 15(3):358–366
Zou X (2005) Studies on inheritance of main quantitative characters and relative mechanism of male sterility in Capsicum. Doctorate dissertation, Nanjing Agricultural University (In Chinese)
Acknowledgments
We thank Dr Zonglie Hong from University of Idaho for critical reading of the manuscript. This work was supported by a grant from the KIC Hubei (No. 2009BFA005), ISTC MOST (No. 2011DFB31620) and SFYS HAAS (No. 2012NKYJJ04).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yao, M., Li, N., Wang, F. et al. Genetic analysis and identification of QTLs for resistance to cucumber mosaic virus in chili pepper (Capsicum annuum L.). Euphytica 193, 135–145 (2013). https://doi.org/10.1007/s10681-013-0953-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-013-0953-8