Abstract
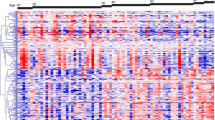
MicroRNAs are non-coding RNAs with roles in many cellular processes. Tissue-specific miRNA profiles associated with senescence have been described for several cell and tissue types. We aimed to characterise miRNAs involved in core, rather than tissue-specific, senescence pathways by assessment of common miRNA expression differences in two different cell types, with follow-up of predicted targets in human peripheral blood. MicroRNAs were profiled in early and late passage primary lung and skin fibroblasts to identify commonly-deregulated miRNAs. Expression changes of their bioinformatically-predicted mRNA targets were then assessed in both cell types and in human peripheral blood from elderly participants in the InCHIANTI study. 57/178 and 26/492 microRNAs were altered in late passage skin and lung cells respectively. Three miRNAs (miR-92a, miR-15b and miR-125a-3p) were altered in both tissues. 14 mRNA targets of the common miRNAs were expressed in lung and skin fibroblasts, of which two demonstrated up-regulation in late passage skin and lung cells (LYST; p = 0.02 [skin] and 0.02 [lung] INMT; p = 0.03 [skin] and 0.04 [lung]). ZMPSTE24 and LHFPL2 demonstrated altered expression in late passage skin cells only (p = 0.01 and 0.05 respectively). LHFPL2 was also positively correlated with age in peripheral blood (p value = 6.6 × 10−5). We find that the majority of senescence-associated miRNAs demonstrate tissue-specific effects. However, miRNAs showing common effects across tissue types may represent those associated with core, rather than tissue-specific senescence processes.

Similar content being viewed by others
References
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Boehm M, Slack F (2005) A developmental timing microRNA and its target regulate life span in C. elegans. Science 310:1954–1957
Brandenberger R et al (2004) Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat Biotechnol 22:707–716
Breitbart RE, Andreadis A, Nadal-Ginard B (1987) Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem 56:467–495
Butler RN et al (2008) New model of health promotion and disease prevention for the 21st century. BMJ 337:149
Denecke J et al (2006) A homozygous ZMPSTE24 null mutation in combination with a heterozygous mutation in the LMNA gene causes Hutchinson-Gilford progeria syndrome (HGPS): insights into the pathophysiology of HGPS. Hum Mutat 27:524–531
Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk—database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inf 44:839–847
Ferrucci L, Bandinelli S, Benvenuti E, Di Iorio A, Macchi C, Harris TB, Guralnik JM (2000) Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc 48:1618–1625
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114
Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315
Ghosh S, Zhou Z (2014) Genetics of aging, progeria and lamin disorders. Curr Opin Genet Dev 26C:41–46
Glass JD, McClusky ME (1987) Immunoreactive luteinizing hormone-containing neurons in the brain of the white-footed mouse, Peromyscus leucopus. Experientia 43:188–190
Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123:631–640
Grillari J, Hackl M, Grillari-Voglauer R (2010) miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology 11:501–506
Hackl M et al (2010) miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell 9:291–296
Harries LW et al (2011) Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell 10:868–878
Harries LW et al (2012a) Leukocyte CCR2 expression is associated with mini-mental state examination score in older adults. Rejuvenation Res 15:395–404
Harries LW et al (2012b) Advancing age is associated with gene expression changes resembling mTOR inhibition: evidence from two human populations. Mech Ageing Dev 133:556–562
Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM (2006) Cellular senescence in aging primates. Science 311:1257
Holly AC, Melzer D, Pilling LC, Fellows AC, Tanaka T, Ferrucci L, Harries LW (2013) Changes in splicing factor expression are associated with advancing age in man. Mech Ageing Dev 134(9):356–366
Inukai S, Slack F (2013) MicroRNAs and the genetic network in aging. J Mol Biol 425(19):3601–3608
Jaenisch R, Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33:245–254
Jeyapalan JC, Sedivy JM (2008) Cellular senescence and organismal aging. Mech Ageing Dev 129:467–474
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:D152–D157
Lal A et al (2008) p16INK4a translation suppressed by miR-24. PLoS One 3:e1864
Lim LP et al (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773
Liu N et al (2012) The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 482:519–523
Longo-Guess CM, Gagnon LH, Cook SA, Wu J, Zheng QY, Johnson KR (2005) A missense mutation in the previously undescribed gene Tmhs underlies deafness in hurry-scurry (hscy) mice. Proc Natl Acad Sci USA 102:7894–7899
Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G (2013) The hallmarks of aging. Cell 153:1194–1217
Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J (2009) A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 10:R64
Nishino J, Kim I, Chada K, Morrison SJ (2008) Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135:227–239
Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK (2010) microRNA expression patterns reveal differential expression of target genes with age. PLoS One 5:e10724
Papackova Z, Cahova M (2014) Important role of autophagy in regulation of metabolic processes in health, disease and aging. Physiol Res 63(4):409–420
Passos JF et al (2007) Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol 5:e110
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 4 29:e45
R-Core-team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing http://www.R-project.org/
Roy A, Kar R, Basu D, Srivani S, Badhe BA (2011) Clinico-hematological profile of Chediak-Higashi syndrome: experience from a tertiary care center in south India. Indian J Pathol Microbiol 54:547–551
Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3(1):1–25
Soifer HS, Rossi JJ, Sætrom P (2007) MicroRNAs in disease and potential therapeutic applications. Mol Ther 15:2070–2079
Soreghan BA et al (2005) Using proteomics and network analysis to elucidate the consequences of synaptic protein oxidation in a PS1+AbetaPP mouse model of Alzheimer’s disease. J Alzheimers Dis 8:227–241
Stefani G, Slack FJ (2008) Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9:219–230
Wang L, Lyerla T (2010) Histochemical and cellular changes accompanying the appearance of lung fibrosis in an experimental mouse model for Hermansky Pudlak syndrome. Histochem Cell Biol 134:205–213
Weindruch R, Kayo T, Lee CK, Prolla TA (2002) Gene expression profiling of aging using DNA microarrays. Mech Ageing Dev 123:177–193
Wood S (2011) Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J Roy Stat Soc 73:3–36
Zeller T et al (2010) Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One 5:e10693
Acknowledgments
The authors would like to acknowledge Dr Jonathan Locke for help and advice regarding the miRNA analysis and Mr Ben Lee for technical assistance. This work was supported internal funds from the University of Exeter Medical School. TvZ acknowledges funding from BBSRC Grant reference BB/I020748/1. SNG acknowledges funding from the Addison Wheeler Trust, Durham University. PvDW was supported by an Erasmus fellowship.
Conflict of interest
The authors report no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Alice C. Holly and Sushma Grellscheid have contributed equally to this publication.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Holly, A.C., Grellscheid, S., van de Walle, P. et al. Comparison of senescence-associated miRNAs in primary skin and lung fibroblasts. Biogerontology 16, 423–434 (2015). https://doi.org/10.1007/s10522-015-9560-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10522-015-9560-5