Abstract
Background and aims
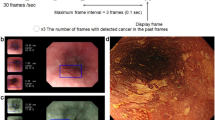
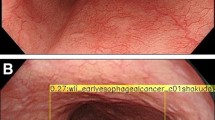
The endocytoscopic system (ECS) helps in virtual realization of histology and can aid in confirming histological diagnosis in vivo. We propose replacing biopsy-based histology for esophageal squamous cell carcinoma (ESCC) by using the ECS. We applied deep-learning artificial intelligence (AI) to analyse ECS images of the esophagus to determine whether AI can support endoscopists for the replacement of biopsy-based histology.
Methods
A convolutional neural network-based AI was constructed based on GoogLeNet and trained using 4715 ECS images of the esophagus (1141 malignant and 3574 non-malignant images). To evaluate the diagnostic accuracy of the AI, an independent test set of 1520 ECS images, collected from 55 consecutive patients (27 ESCCs and 28 benign esophageal lesions) were examined.
Results
On the basis of the receiver-operating characteristic curve analysis, the areas under the curve of the total images, higher magnification pictures, and lower magnification pictures were 0.85, 0.90, and 0.72, respectively. The AI correctly diagnosed 25 of the 27 ESCC cases, with an overall sensitivity of 92.6%. Twenty-five of the 28 non-cancerous lesions were diagnosed as non-malignant, with a specificity of 89.3% and an overall accuracy of 90.9%. Two cases of malignant lesions, misdiagnosed as non-malignant by the AI, were correctly diagnosed as malignant by the endoscopist. Among the 3 cases of non-cancerous lesions diagnosed as malignant by the AI, 2 were of radiation-related esophagitis and one was of gastroesophageal reflux disease.
Conclusion
AI is expected to support endoscopists in diagnosing ESCC based on ECS images without biopsy-based histological reference.



Similar content being viewed by others
References
Kumagai Y, Monma K, Kawada K. Magnifying chromoendoscopy of the esophagus: in vivo pathological diagnosis using an endocytoscopy system. Endoscopy. 2004;36:590–4.
Kumagai Y, Kawada K, Yamazaki S, et al. Endocytoscopic observation for esophageal squamous cell carcinoma: can biopsy histology be omitted? Dis Esophagus. 2009;22:505–12.
Kumagai Y, Kawada K, Yamazaki S, et al. Prospective replacement of magnifying endoscopy by a newly developed endocytoscope, the ‘GIF-Y0002’. Dis Esophagus. 2010;23:627–32.
Kumagai Y, Kawada K, Yamazaki S, et al. Current status and limitations of the newly developed endocytoscope GIF-Y0002 with reference to its diagnostic performance for common esophageal lesions. J Dig Dis. 2012;13:393–400.
Kumagai Y, Kawada K, Yamazaki S, et al. Endocytoscopic observation of esophageal squamous cell carcinoma. Dig Endosc. 2010;22:10–6.
Kumagai Y, Kawada K, Higashi M, et al. Endocytoscopic observation of various esophageal lesions at ×600: can nuclear abnormality be recognized? Dis Esophagus. 2015;28:269–75.
Kumagai Y, Takubo K, Kawada K, et al. Endocytoscpic observation of various types of esophagitis. Esophagus. 2016;13:200–7. https://doi.org/10.1007/s10388-015-0517-1.
Kumagai Y, Takubo K, Kawada K, et al. A newly developed continuous zoom-focus endocytoscope. Endoscopy. 2017;49(2):176–80.
Inoue H, Kazawa T, Sato Y, et al. In vivo observation of living cancer cells in the esophagus, stomach, and colon using catheter-type contact endoscope, “Endo-Cytoscopy system”. Gastrointest Endosc Clin N Am. 2004;14:589–94.
Sasajima K, Kudo SE, Inoue H, et al. Real-time in vivo virtual histology of colorectal lesions when using the endocytoscopy system. Gastrointest Endosc. 2006;63(7):1010–7.
Bibault JE, Giraud P, Burgun A. Big Data and machine learning in radiation oncology: state of the art and future prospects. Cancer Lett. 2016;382:110–7.
Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–8.
Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–10.
Yoshida H, Shimazu T, Kiyuna T, et al. Automated histological classification of whole-slide images of gastric biopsy specimens. Gastric Cancer. 2018;21(2):249–57. https://doi.org/10.1007/s10120-017-0731-8.
Misawa M, Kudo S, Mori Y, et al. Accuracy of computer-aided diagnosis based on narrow-band imaging endocytoscopy for diagnosing colorectal lesions: comparison with experts. Int J Comput Assist Radiol Surg. 2017;12:757–66.
Shichijo S, Nomura S, Aoyama K, et al. Application of convolutional neural networks in the diagnosis of Helicobacter pylori infection based on endoscopic images. EBioMedicine. 2017;25:106–11.
Hirasawa T, Aoyama K, Tanimoto T, et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21(4):653–60. https://doi.org/10.1007/s10120-018-0793-2.
Inoue H, Kazawa T, Sato Y, et al. In vivo observation of living cancer cells in the esophagus, stomach, and colon using catheter-type contact endoscope, “Endo-Cytoscopy system”. Gastrointest Endosc Clin N Am. 2004;14:589–94.
Misawa M, Kudo S, Mori Y, et al. Artificial intelligence-assisted polyp detection for colonoscopy: initial experience. Gastroenterology. 2018;154(8):2027.e3–2029.e3. https://doi.org/10.1053/j.gastro.2018.04.003.
Takubo K. Pathology of the esophagus. 3rd ed. Tokyo: Wiley Publishing Japan; 2017. p. 88–102 (131–134 ).
Acknowledgements
We are grateful to Tomoyuki Kawada for his kind advice and confirmation of statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
T. Tada, Y. Endo and K. Aoyama are employed by AI Medical Service Inc. The other authors declare no conflict of interest.
Human rights statement
All procedures followed were in accordance with the ethical standards of our hospital ethics committee on human experimentation and with the Helsinki Declaration of 1964 and later versions.
Informed consent
Written informed consent for endocytoscopic observation was obtained from all patients. Opt out consent was obtained for AI analysis.
Rights and permissions
About this article
Cite this article
Kumagai, Y., Takubo, K., Kawada, K. et al. Diagnosis using deep-learning artificial intelligence based on the endocytoscopic observation of the esophagus. Esophagus 16, 180–187 (2019). https://doi.org/10.1007/s10388-018-0651-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10388-018-0651-7