Abstract
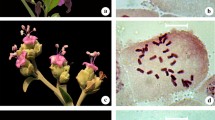
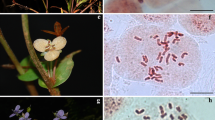
Chromosome numbers and karyotypes of 26 Ophiopogon species, 2 Liriope species and 5 Peliosanthes species of the family Liliaceae from Southwest China, were investigated. The study revealed a detailed picture of chromosome features and their pattern of karyotype variation in Ophiopogoneae. Karyotype asymmetry in different species and different populations of the same species varied greatly due to different locality conditions. Our analyses may support the separately monophyly of Ophiopogon, Liriope and Peliosanthes.




Similar content being viewed by others
References
Bailey LH (1929) The case of Ophiopogon and Liriope. Gentes Herb 2:3–37
Brandham PE, Doherty MJ (1998) Genome size variation in the Aloaceae, an angiosperm family displaying karyotypic orthoselection. Ann Bot 82:67–73
Chen XQ, Tamura MN (2000) Ophiopogon Ker-Gawl. FOC 24:252–261
Cutler DF (1992) Vegetative anatomy of Ophiopogoneae (Convallariaceae). Bot J Linn Soc 110:385–419
Dai LK, Liang SJ (1991) Epidermal features of leaves and their taxonomic signification in subfamily Ophiopogonoideae (Liliaceae). Acta Phytotaxon Sin 29:335–346
Hume HH (1961) The Liriope-Ophiopogon complex. Baileya 46:135–158
Kim JH, Kim DK, Foreat F, Fay MF, Chase MW (2010) Molecular phylogenetics of Ruscaceae sense lato and related families (Asparagales) based on plastid and nuclear DNA sequences. Ann Bot 106:775–790
Levan A, Fedga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220
Liang SJ, Dai LK (1992) Pollen morphology and generic phylogenetic relationships in Ophiopogonoideae (Liliaceae). Acta Phytotaxon Sin 30:427–437
Malik CP (1962) Cytological studies on the three Indian species of Ophiopogon. Phyton 19:35–42
Martel E, Poncet V, Lamy F, Siljak-Yakovlev S, Lejeune B, Sarr A (2004) Chromosome evolution of Pennisetum species (Poaceae): implications of ITS phylogeny. Plant Syst Evol 249:139–149
Mcharo M, Bush E, Bonte DL, Broussard C, Urbatsch L (2003) Molecular and morphological investigation of ornamental liriopogons. J Am Soc for Hortic Sci 128:575–577
Nagamatsu T, Noda S (1971) Balanced hypotetraploids in Ophiopogon japonicus and O. ohwii. Cytologia 36:332–340
Rudall PJ, Gasjon P (2000) Systematics of Ruscaceae/Convallariaceae: a combined morphological and molecular investigation. Bot J Linn Soc 134:73–92
Sarker AK, Datta R, Raychowdhury M (1974) IOPB chromosome number reports XLVI. Taxon 23:801–812
Sharma AK (1970) Annual report, 1967–1968. Res Bull Univ Calcutta 2:1–50
Sharma AK, Chandhnri M (1964) Cytological studies as an aid in assessing the status of Sansevieria, Ophiopogon and Curculigo. Nucleus 7:43–58
Skinner HT (1971) Some liriopogon comments. J Roy Hort Soc 96:345–350
Stebbins GL (1971) Chromosomal evolution in higher plants. Edward Arnold, London
Tamura MN, Yamashita J (2004) Molecular phylogeny of monocotyledons inferred from combined analysis of plastid matK and rbcL gene sequences. J Plant Res 117:109–120
Tanaka R (1971) Types of resting nuclei in Orchidaceae. Bot Mag Tokyo 84:118–122
Tanaka R (1977) Recent karyotype studies. In: Ogawa K, Koike S, Kurosumi I, SatoMeds. Plant cytology. Asakura Publisher, Tokyo, pp 293–326
Tanaka R (1987) The karyotype theory andwide crossing as an example in Orchidaceae. In: Hong DY (ed) Plant chromosome research 1989. Proc Sino-Japanese Symp Plant Chromosomes, Hiroshima, pp 1–10
Yamashita J, Tamura MN (2001) Karyotype analysis of six species of the genus Ophiopogon (Convallariaceae-Ophiopogoneae). Jap J Bot 76:100–119
Yang YP, Li H, Liu XZ, Kondo K (1990) Karyotype study of the genus Ophiopogon in Yunnan. Acta Bot Yunnan (Suppl 3):94–102
Zhang DM (1991) Chromosomal study and an insight into systematics of the tribe Ophiopogoneae (Endl.) Kunth. Ph.D. Dissertation, Inst Bot, Chinese Acad Sci, Beijing
Acknowledgments
We are grateful to Professor Jan Suda for his important assistance in manuscript revision, and Dr. Hu Guangwan for providing some necessary materials. The study was supported by grants from the Ministry of Science and Technology of China, Major State Basic Research Development Program (2010CB951700), and the National Natural Science Foundation of China (NSFC 40930209 to H. Sun).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, GY., Meng, Y. & Yang, YP. Karyological analyses of 33 species of the tribe Ophiopogoneae (Liliaceae) from Southwest China. J Plant Res 126, 597–604 (2013). https://doi.org/10.1007/s10265-013-0557-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-013-0557-3