Abstract
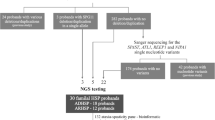
Mutations in the spatacsin gene have recently been identified as the genetic cause of autosomal–recessive spastic paraplegia (SPG) with thin corpus callosum, mapping to chromosome 15p13–21. While several nonsense and frameshift mutations as well as splice mutations have been identified, large genomic deletions have not yet been found, potentially due to the absence of an efficient analysis tool. After complete sequencing of 12 autosomal recessive hereditary spastic paraplegia patients with suggestive clinical signs, we were able to define nine SPG11 cases but were left with three patients in which only one SPG11 mutation could be identified by direct sequencing. In these patients, we performed high-resolution comparative genomic hybridization using a predesigned human chromosome 15 tiling array with an average spacing of 100 bp. Data analysis suggested heterozygous genomic deletion within the spatacsin gene in all three patients. In one patient, a relatively small genomic deletion (8.2 kb) could be validated by quantitative polymerase chain reaction (PCR) and long-range PCR, allowing the diagnosis of the deletion of exons 31 through 34. For two patients, quantitative PCR validation could not confirm a genomic deletion. As high density tiling arrays are available for the entire human genome, we suggest this approach for the screening of heterozygous genomic deletions in candidate genes down to a few kilobases.


Similar content being viewed by others
References
Stevanin G, Santorelli FM, Azzedine H, Coutinho P, Chomilier J, Denora PS et al (2007) Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nat Genet 39(3):366–372, doi:10.1038/ng1980
Hehr U, Bauer P, Winner B, Schule R, Olmez A, Koehler W et al (2007) Long-term course and mutational spectrum of spatacsin-linked spastic paraplegia. Ann Neurol 62(6):656–665, doi:10.1002/ana.21310
Stevanin G, Azzedine H, Denora P, Boukhris A, Tazir M, Lossos A, Rosa AL, Lerer I, Hamri A, Alegria P, Loureiro J, Tada M, Hannequin D, Anheim M, Goizet C, Gonzalez-Martinez V, Le Ber I, , Forlani S, Iwabuchi K, Meiner V, Uyanik G, Erichsen AK, Feki I, Pasquier F, Belarbi S, Cruz VT, Depienne C, Truchetto J, Garrigues G, Tallaksen C, Tranchant C, Nishizawa M, Vale J, Coutinho P, Santorelli FM, Mhiri C, Brice A, Durr A (2008) Mutations in SPG11 are frequent in autosomal recessive spastic paraplegia with thin corpus callosum, cognitive decline and lower motor neuron degeneration. Brain 131(Pt 3):772–784, doi:10.1093/brain/awm293
Del Bo R, Di Fonzo A, Ghezzi S, Locatelli F, Stevanin G, Costa A et al (2007) SPG11: a consistent clinical phenotype in a family with homozygous Spatacsin truncating mutation. Neurogenetics 8(4):301–305, doi:10.1007/s10048-007-0095-z
Lee MJ, Cheng TW, Hua MS, Pan MK, Wang J, Stephenson DA et al (2008) Mutations of the SPG11 gene in patients with autosomal recessive spastic paraparesis and thin corpus callosum. J Neurol Neurosurg Psychiatry 79(5):607–609, doi:10.1136/jnnp.2007.136390
Zhang SS, Chen Q, Chen XP, Wang JG, Burgunder JM, Shang HF et al (2008) Two novel mutations in the SPG11 gene causing hereditary spastic paraplegia associated with thin corpus callosum. Mov Disord 23(6):917–919, doi:10.1002/mds.21942
Paisan-Ruiz C, Dogu O, Yilmaz A, Houlden H, Singleton A (2008) SPG11 mutations are common in familial cases of complicated hereditary spastic paraplegia. Neurology 70(16 Pt 2):1384–1389, doi:10.1212/01.wnl.0000294327.66106.3d
Boukhris A, Stevanin G, Feki I, Denis E, Elleuch N, Miladi MI et al (2008) Hereditary spastic paraplegia with mental impairment and thin corpus callosum in Tunisia: SPG11, SPG15, and further genetic heterogeneity. Arch Neurol 65(3):393–402, doi:10.1001/archneur.65.3.393
Simon-Sanchez J, Scholz S, Matarin MM, Fung HC, Hernandez D, Gibbs JR et al (2008) Genomewide SNP assay reveals mutations underlying Parkinson disease. Human Mutat 29(2):315–322, doi:10.1002/humu.20626
Beetz C, Nygren AO, Schickel J, Auer-Grumbach M, Burk K, Heide G, Kassubek J, Klimpe S, Klopstock T, Kreuz F, Otto S, Schule R, Schols L, Sperfeld AD, Witte OW, Deufel T (2006) High frequency of partial SPAST deletions in autosomal dominant hereditary spastic paraplegia. Neurology 67(11):1926–1930, doi:10.1212/01.wnl.0000244413.49258.f5
Depienne C, Fedirko E, Forlani S, Cazeneuve C, Ribai P, Feki I et al (2007) Exon deletions of SPG4 are a frequent cause of hereditary spastic paraplegia. J Med Genet 44(4):281–284, doi:10.1136/jmg.2006.046425
Janssen B, Hartmann C, Scholz V, Jauch A, Zschocke J (2005) MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics 6(1):29–35, doi:10.1007/s10048-004-0204-1
Scarciolla O, Brancati F, Valente EM, Ferraris A, De Angelis MV, Valbonesi S et al (2007) Multiplex ligation-dependent probe amplification assay for simultaneous detection of Parkinson’s disease gene rearrangements. Mov Disord 22(15):2274–2278, doi:10.1002/mds.21532
Slater H, Bruno D, Ren H, La P, Burgess T, Hills L et al (2004) Improved testing for CMT1A and HNPP using multiplex ligation-dependent probe amplification (MLPA) with rapid DNA preparations: comparison with the interphase FISH method. Human Mutat 24(2):164–171, doi:10.1002/humu.20072
Sellner LN, Taylor GR (2004) MLPA and MAPH: new techniques for detection of gene deletions. Human Mutat 23(5):413–419, doi:10.1002/humu.20035
Laccone F, Junemann I, Whatley S, Morgan R, Butler R, Huppke P et al (2004) Large deletions of the MECP2 gene detected by gene dosage analysis in patients with Rett syndrome. Human Mutat 23(3):234–244, doi:10.1002/humu.20004
Shadrina MI, Semenova EV, Slominsky PA, Bagyeva GH, Illarioshkin SN, Ivanova-Smolenskaia II et al (2007) Effective quantitative real-time polymerase chain reaction analysis of the parkin gene (PARK2) exon 1–12 dosage. BMC Med Genet 8:6, doi:10.1186/1471-2350-8-6
Urban AE, Korbel JO, Selzer R, Richmond T, Hacker A, Popescu GV et al (2006) High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc Natl Acad Sci U S A 103(12):4534–4539, doi:10.1073/pnas.0511340103
Lissens W, Sermon K (1997) Preimplantation genetic diagnosis: current status and new developments. Hum Reprod 12(8):1756–1761, doi:10.1093/humrep/12.8.1756
Mefford HC, Clauin S, Sharp AJ, Moller RS, Ullmann R, Kapur R (2007) Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet 81(5):1057–1069, doi:10.1086/522591
Winner B, Uyanik G, Gross C, Lange M, Schulte-Mattler W, Schuierer G et al (2004) Clinical progression and genetic analysis in hereditary spastic paraplegia with thin corpus callosum in spastic gait gene 11 (SPG11). Arch Neurol 61(1):117–121, doi:10.1001/archneur.61.1.117
Schule R, Holland-Letz T, Klimpe S, Kassubek J, Klopstock T, Mall V et al (2006) The Spastic Paraplegia Rating Scale (SPRS): a reliable and valid measure of disease severity. Neurology 67(3):430–434, doi:10.1212/01.wnl.0000228242.53336.90
Acknowledgements
P.B., R.S., O.R., and L.S. have the support of the German Ministry of Education and Research through funding for the German Network for Movement Disorders (GeNeMove); grant 01GM0603. U.H, B.W., and the e-rare consortium EUROSPA (European and Mediterranean network on spastic paraplegias) J.W. received support from the Tom-Wahlig-Stiftung e.V.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Rights and permissions
About this article
Cite this article
Bauer, P., Winner, B., Schüle, R. et al. Identification of a heterozygous genomic deletion in the spatacsin gene in SPG11 patients using high-resolution comparative genomic hybridization. Neurogenetics 10, 43–48 (2009). https://doi.org/10.1007/s10048-008-0144-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-008-0144-2