Abstract
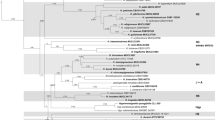
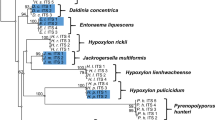
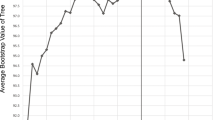
The internal transcribed spacer region (ITS) of the nuclear rDNA cistron represents the barcoding locus for Fungi. Intragenomic variation of this multicopy gene can interfere with accurate phylogenetic reconstruction of biological entities. We investigated the amount and nature of this variation for the lichenized fungus Cora inversa in the Hygrophoraceae (Basidiomycota: Agaricales), analyzing base call and length variation in ITS1 454 pyrosequencing data of three samples of the target mycobiont, for a total of 16,665 reads obtained from three separate repeats of the same samples under different conditions. Using multiple fixed alignment methods (PaPaRa) and maximum likelihood phylogenetic analysis (RAxML), we assessed phylogenetic relationships of the obtained reads, together with Sanger ITS sequences from the same samples. Phylogenetic analysis showed that all ITS1 reads belonged to a single species, C. inversa. Pyrosequencing data showed 266 insertion sites in addition to the 325 sites expected from Sanger sequences, for a total of 15,654 insertions (0.94 insertions per read). An additional 3,279 substitutions relative to the Sanger sequences were detected in the dataset, out of 5,461,125 bases to be called. Up to 99.3 % of the observed indels in the dataset could be interpreted as 454 pyrosequencing errors, approximately 65 % corresponding to incorrectly recovered homopolymer segments, and 35 % to carry-forward-incomplete-extension errors. Comparison of automated clustering and alignment-based phylogenetic analysis demonstrated that clustering of these reads produced a 35-fold overestimation of biological diversity in the dataset at the 95 % similarity threshold level, whereas phylogenetic analysis using a maximum likelihood approach accurately recovered a single biological entity. We conclude that variation detected in 454 pyrosequencing data must be interpreted with great care and that a combination of a sufficiently large number of reads per taxon, a set of Sanger references for the same taxon, and at least two runs under different emulsion PCR and sequencing conditions, are necessary to reliably separate biological variation from 454 sequencing errors. Our study shows that clustering methods are highly sensitive to artifactual sequence variation and inadequate to properly recover biological diversity in a dataset, if sequencing errors are substantial and not removed prior to clustering analysis.


Similar content being viewed by others
References
454 Life Sciences Corporation (2009) Genome sequencer FLX system software manual, version 2.3. 454 Life Sciences Corp., A Roche Company, Brandford, CT
Abe T, Ikemura T, Sugahara J, Kanai A, Ohara Y, Uehara H, Kinouchi M, Kanaya S, Yamada Y, Muto A, Inokuchi I (2011) tRNADB-CE 2011: tRNA gene database curated manually by experts. Nucleic Acids Res 39:D210–D213
Ahmadian A, Ehn M, Hober S (2006) Pyrosequencing: history, biochemistry and future. Clin Chim Acta 363:83–94
Amend A, Samson R, Seifert K, Bruns T (2010) Deep sequencing reveals diverse and geographically structured assemblages of fungi in indoor dust. Proc Natl Acad Sci USA 107:13748–13753
Balzer S, Malde K, Jonassen I (2011) Systematic exploration of error sources in pyrosequencing flowgram data. Bioinformatics 27:304–309
Begerow D, Nilsson H, Unterseher M, Maier W (2010) Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl Microbiol Biotechnol 87:99–108
Berger SA, Stamatakis A (2011) Aligning short reads to reference alignments and trees. Bioinformatics 27:2068–2075
Berger SA, Krompass D, Stamatakis A (2011) Performance, accuracy and web-server for evolutionary placement of short sequence reads under maximum-likelihood. Syst Biol 60:291–302
Borsuk P, Gniadkowski M, Bartnik E, Stepien PP (1988) Unusual evolutionary conservation of 5S rRNA pseudogenes in Aspergillus nidulans: similarity of the DNA sequence associated with the pseudogenes with the mouse immunoglobulin switch region. J Mol Evol 28:125–130
Buée M, Reich M, Murat C, Nilsson RH, Uroz S, Martin F (2009) 454 Pyrosequencing analyzes of forest soils reveal an unexpectedly high fungal diversity. New Phytol 184:449–456
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010a) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010b) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
Dai M, Hamel C, Arnaud M, He Y, Grant C, Lupwayi N, Janzen H, Malhi SS, Yang X, Zhou Z (2012) Arbuscular mycorrhizal fungi assemblages in Chernozem great groups revealed by massively parallel pyrosequencing. Can J Microbiol 58:81–92
Dal-Forno M, Lawrey JD, Sikaroodi M, Bhattarai S, Gillevet PM, Sulzbacher M, Lücking R (2013) Starting from scratch: evolution of the lichen thallus in the basidiolichen Dictyonema (Agaricales: Hygrophoraceae). Fungal Biol 117(9):584–598
Del-Prado R, Cubas P, Lumbsch HT, Divakar PK, Blanco O, de Paz GA, Molina MC, Crespo A (2010) Genetic distances within and among species in monophyletic lineages of Parmeliaceae (Ascomycota) as a tool for taxon delimitation. Mol Phylogenet Evol 56:125–133
Deng W, Maust BS, Westfall DH, Chen L, Zhao H, Larsen BB, Iyer S, Liu Y, Mullins JI (2013) Indel and carryforward correction (ICC): a new analysis approach for processing 454 pyrosequencing data. Bioinformatics 29(19):2402–2409. doi:10.1093/bioinformatics/btt434
Eberhardt U (2010) A constructive step towards selecting a DNA barcode for fungi. New Phytol 187:265–268
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996–998. doi:10.1038/nmeth.2604
Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P (2010) Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J 4:642–647
Ganley ARD, Kobayashi T (2007) Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res 17:184–191
Ganley ARD, Kobayashi T (2011) Monitoring the rate and dynamics of concerted evolution in the ribosomal DNA repeats of Saccharomyces cerevisiae using experimental evolution. Mol Biol Evol 28:2883–2891
Ganley AR, Scott B (2002) Concerted evolution in the ribosomal RNA genes of an Epichloë endophyte hybrid: comparison between tandemly arranged rDNA and dispersed 5S rrn genes. Fungal Genet Biol 35:39–51
Geml J, Laursen GA, Taylor DL (2008) Molecular diversity assessment of arctic and boreal Agaricus taxa. Mycologia 100:577–589
Gilles A, Meglécz E, Pech N, Ferreira S, Malausa T, Martin JF (2011) Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics 12:245
Gomes EA, Kasuya MCM, de Barros EG, Borges AC, Araujo EF (2002) Polymorphism in the internal transcribed spacer (ITS) of the ribosomal DNA of 26 isolates of ectomycorrhizal fungi. Genet Mol Biol 25:477–483
Gomez-Alvarez V, Teal TK, Schmidt TM (2009) Systematic artifacts in metagenomes from complex microbial communities. ISME J 3:1314–1317
Gulyas G, Sramko G, Molnar A, Rudnoy S, Illyes Z, Balazs T, Zoltan B (2005) Nuclear ribosomal DNA ITS paralogs as evidence of recent hybridization in genus Ophrys (Orchidaceae). Acta Biol Crac Ser Bot 47:61–67
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hebert PD, Ratnasingham S, deWaard JR (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci 270(Suppl 1):S96–S99
Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S, Zody MC, Erlich RL, Green LM, Berical A, Wang Y, Casali M, Steeck H, Bloom AK, Dudek T, Tully D, Newman R, Axten KL, Gladden AD, Battis L, Kemper M, Zeng Q, Shea TP, Gujja S, Zedlack C, Gasser O, Brander C, Hess C, Gunthard HF, Brumme ZL, Brumme CJ, Bazner S, Rychert J, Tinsley JP, Mayer KH, Rosenberg E, Pereya F, Levin JZ, Young SK, Jessen H, Altfeld M, Birren BW, Walker BD, Allen TM (2012) Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog 8(3):e1002529
Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk PM, Nilsson RH (2011) Progress in molecular and morphological taxon discovery in fungi and options for formal classification of environmental sequences. Fungal Biol Rev 25:38–47
Hoff KJ (2009) The effect of sequencing errors on metagenomic gene prediction. BMC Genomics 10:520
Huang Y, Niu B, Gao Y, Fu L, Li W (2010) CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680
Hughes KW, Petersen RH (2001) Apparent recombination or gene conversion in the ribosomal ITS region of a Flammulina (Fungi, Agaricales) hybrid. Mol Biol Evol 18:94–96
Hurst LD, Smith NGC (1998) The evolution of concerted evolution. Proc Biol Sci 265:121–127
Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8:R143
Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898
James SA, O’Kelly MJ, Carter DM, Davey RP, van Oudenaarden A, Roberts IN (2009) Repetitive sequence variation and dynamics in the ribosomal DNA array of Saccharomyces cerevisiae as revealed by whole-genome resequencing. Genome Res 19:626–635
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res 30:3059–3066
Katoh K, Asimenos G, Toh H (2009) Multiple alignment of DNA sequences with MAFFT. Meth Mol Biol 537:39–64
Kauserud H, Kumar S, Brysting A, Nordén J, Carlsen T (2012) High consistency between replicate 454 pyrosequencing analyses of ectomycorrhizal plant root samples. Mycorrhiza 22:309–315
Keegan KP, Trimble WL, Wilkening J, Wilke A, Harrison T, D’Souza M, Meyer F (2012) A platform-independent method for detecting errors in metagenomic sequencing data: dRISEE. PLoS Comput Biol 8(6):e1002541
Kiss L (2012) Limits of nuclear ribosomal DNA internal transcribed spacer (ITS) sequences as species barcodes for Fungi. Proc Natl Acad Sci USA 109:E1811
Ko KS, Jung HS (2002) Three nonorthologous ITS1 types are present in a polypore fungus Trichaptum abietinum. Mol Phylogenet Evol 23:112–122
Kõljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AF, Tedersoo L, Vrålstad T, Ursing BM (2005) UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol 166:1063–1068
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny B, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Oldmaa KP, Saag L, Saar R, Schüssler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. doi:10.1111/mec.12481
Kovács GM, Balázs TK, Calonge FD, Martín MP (2011a) The diversity of Terfezia desert truffles: new species and a highly variable species complex with intrasporocarpic nrDNA ITS heterogeneity. Mycologia 103:841–853
Kovács GM, Jankovics T, Kiss L (2011b) Variation in the nrDNA ITS sequences of some powdery mildew species: do routine molecular identification procedures hide valuable information? Eur J Plant Pathol 131:135–141
Kumar S, Carlsen T, Mevik B, Enger P, Blaalid R, Shalchian-Tabrizi K, Kauserud H (2011) CLOTU: an online pipeline for processing and clustering of 454 amplicon reads into OTUs followed by taxonomic annotation. BMC Bioinformatics 12:82
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123
Lawrey JD, Lücking R, Sipman HJM, Chaves JL, Redhead SA, Bungartz F, Sikaroodi M, Gillevet PM (2009) High concentration of basidiolichens in a single family of agaricoid mushrooms. Mycol Res 113:1154–1171
Li W, Godzik A (2006) Cd-Hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Li Y, Jiao L, Yao YJ (2013) Non-concerted ITS evolution in fungi, as revealed from the important medicinal fungus Ophiocordyceps sinensis. Mol Phylogen Evol 68(2):373–379
Liao D (1999) Concerted evolution: molecular mechanism and biological implications. Am J Hum Genet 64:24–30
Liao D (2008) Concerted evolution. University of Florida, Gainesville, FL. doi:10.1002/9780470015902.a0005132
Lindner DL, Banik MT (2011) Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and inflates estimates of operational taxonomic units in genus Laetiporus. Mycologia 103:731–740
Lindner DL, Carlsen T, Nilsson RH, Davey M, Schumacher T, Kauserud H (2013) Employing 454 amplicon pyrosequencing to reveal intragenomic divergence in the internal transcribed spacer (ITS) rDNA region in fungi. Ecol Evol 3:1751–1764
Liu K, Raghavan S, Nelesen S, Linder CR, Warrnow T (2009) Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science 324:1561–1564
Liu K, Warnow TJ, Holder MT, Nelesen S, Yu J, Stamatakis A, Linder CR (2012) SATé-II: very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic Trees. Syst Biol 61:90–106
Lücking R, Kalb K, Essene A (2012) The power of ITS: using megaphylogenies of barcoding genes to reveal inconsistencies in taxonomic identifications of genbank submissions. The 7th IAL symposium “Lichens: from genome to ecosystems in a changing world”, January 2012, Bangkok. Book of abstracts: 3B-1-O2
Lumini E, Orgiazzi A, Borriello R, Bonfante P, Bianciotto V (2010) Disclosing arbuscular mycorrhizal fungal biodiversity in soil through a land-use gradient using a pyrosequencing approach. Environ Microbiol 12:2165–2179
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing inmicrofabricated high-density picolitre reactors. Nature 437:376–380
McGuire KL, Fierer N, Bateman C, Treseder KK, Turner BL (2012) Fungal community composition in neotropical rain forests: the influence of tree diversity and precipitation. Microb Ecol 63:804–812
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE), New Orleans, pp 1–8
Moncada B, Lücking R, Suárez A (2013) Molecular phylogeny of the genus Sticta (lichenized Ascomycota: Lobariaceae) in Colombia. Fungal Div. doi:10.1007/s13225-013-0230-0
Morzycka-Wroblewska E, Selker EU, Stevens JN, Metzenberg RL (1985) Concerted evolution of dispersed Neurospora crassa 5S RNA genes: pattern of sequence conservation between allelic and nonallelic genes. Mol Cell Biol 5:46–51
Nilsson RH, Kristiansson E, Ryberg M, Larsson KH (2005) Approaching the taxonomic affiliation of unidentified sequences in public databases—an example from the mycorrhizal fungi. BMC Bioinformatics 6:178
Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson KH (2006) Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS ONE 1:e59
Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH (2008) Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evolut Bioinformatics 4:193–201
Nilsson RH, Ryberg M, Abarenkov K, Sjökvist E, Kristiansson E (2009) The ITS region as a target for characterization of fungal communities using emerging sequencing technologies. FEMS Microbiol Lett 296:97–101
Nilsson RH, Tedersoo L, Lindahl BD, Kjøller R, Carlsen T, Quince C, Abarenkov K, Pennanen T, Stenlid J, Bruns T, Larsson K-H, Kõljalg U, Kauserud H (2011) Towards standardization of the description and publication of next-generation sequencing datasets of fungal communities. New Phytol 191:314–318
Niu B, Fu L, Sun S, Li W (2010) Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics 11:187
O’Brien HE, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R (2005) Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol 71:5544–5550
Pawlowska TE (2005) Genetic processes in arbuscular mycorrhizal fungi. FEMS Microbiol Lett 251:185–192
Pawlowska TE (2007) How the genome is organized in the Glomeromycota. In: Heitman J, Kronstad JW, Taylor JW, Casselton LA (eds) Sex in Fungi: molecular determination and evolutionary implications. ASM Press, Washington,DC, pp 419–430
Pawlowska TE (2010) Population genetics of fungal mutualists of plants. In: Xu J (ed) Microbial population genetics. Horizon Scientific Press, Norfolk
Pinto PM, Resende MA, Koga-Ito CY, Ferreira JA, Tendler M (2004) rDNA-RFLP identification of Candida species in immunocompromised and seriously diseased patients. Can J Microbiol 50:504–520
Porter TM, Golding GB (2011) Are similarity- or phylogeny-based methods more appropriate for classifying internal transcribed spacer (ITS) metagenomic amplicons? New Phytol 192:775–782
Powell JR, Monaghan MT, Opik M, Rillig MC (2011) Evolutionary criteria outperform operational approaches in producing ecologically relevant fungal species inventories. Mol Ecol 20:655–666
Quince C, Curtis TP, Sloan WT (2008) The rational exploration of microbial diversity. ISME J 2:997–1006
Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6:639–641
Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ (2011) Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38
Reeder J, Knight R (2010) Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7:668–669
Rooney AP, Ward TJ (2005) Evolution of a large ribosomal RNA multigene family in filamentous fungi: birth and death of a concerted evolution paradigm. Proc Natl Acad Sci USA 102:5084–5089
Rossman AY (2007) Report of the planning workshop for all fungi DNA Barcoding. Inoculum 58:1–5
Rothberg JM, Leamon JH (2008) The development and impact of 454 pyrosequencing. Nat Biotechnol 26:1117–1124
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109:6241–6246
Seifert KA (2008) The all-fungi barcoding campaign (FunBOL). Persoonia 20:106
Sikaroodi M, Gillevet PM (2012) Quality control in multi-tag pyrosequencing of microbial communities. Biotechniques 53:381–383
Simon UK, Weiss M (2008) Intragenomic variation of fungal ribosomal genes is higher than previously thought. Mol Biol Evol 25:2251–2254
Smith ME, Douhan GW, Rizzo DM (2007) Intra-specific and intra-sporocarp ITS variation of ectomycorrhizal fungi as assessed by rDNA sequencing of sporocarps and pooled ectomycorrhizal roots from a Quercus woodland. Mycorrhiza 18:15–22
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103:12115–12120
Stamatakis A (2006) RAxML-VI-HPC: maximum-Likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stamatakis A, Ludwig T, Meier H (2005) RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463
Stamatakis A, Hoover P, Rougemont J (2008) A fast bootstrapping algorithm for the RAxML web-servers. Syst Biol 57:758–771
Suchard MA, Redelings BD (2006) BAli-Phy: simultaneous bayesian inference of alignment and phylogeny. Bioinformatics 22:2047–2048
Taylor DL, McCormick MK (2008) Internal transcribed spacer primers and sequences for improved characterization of basidiomycetous orchid mycorrhizas. New Phytol 177:1020–1033
Taylor DL, Booth MG, McFarland JW, Herriott IC, Lennon NJ, Nusbaum C, Marr TG (2008) Increasing ecological infrence from high throughput sequencing of fungi in the environment through a tagging approach. Mol Ecol Resour 8:742–752
Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, Saar I, Bahram M, Bechem E, Chuyong G, Kõljalg U (2010) 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol 188:291–301
Unterseher M, Jumpponen A, Öpik M, Tedersoo L, Moora M, Dormann C, Schnittler M (2011) Species abundance distributions and richness estimations in fungal metagenomics—lessons learned from community ecology. Mol Ecol 20:275–285
Vandenbroucke I, Van Marck H, Verhasselt P, Thys K, Mostmans W, Dumont S, Van Eygen V, Coen K, Tuefferd M, Aerssens J (2011) Minor variant detection in amplicons using 454 massive parallel pyrosequencing: experiences and considerations for successful applications. Biotechniques 51:167–177
Varón A, Vinh LS, Wheeler WC (2010) POY version 4: phylogenetic analysis using dynamic homologies. Cladistics 26:72–85
Wallander H, Johansson U, Sterkenburg E, Durling MB, Lindahl B (2010) Production of ectomycorrhizal mycelium peaks during canopy closure in Norway spruce. New Phytol 187:1124–1134
Wang Z, Nilsson RH, Lopez-Giraldez F, Zhuang WY, Dai YC et al (2011) Tasting soil fungal diversity with earth tongues: phylogenetic test of SATe′ alignments for environmental ITS data. PLoS ONE 6(4):e19039
Won H, Renner SS (2005) The internal transcribed spacer of nuclear ribosomal DNA in the gymnosperm Gnetum. Mol Phylogen Evol 36:581–597
Zhou J, Wu L, Deng Y, Zhi X, Jiang YH, Tu Q, Xie J, Van Nostrand JD, He Z, Yang Y (2011) Reproducibility and quantitation of amplicon sequencing-based detection. ISME J 1:11
Acknowledgments
Financial support for this study was provided by grant DEB 0841405 from the National Science Foundation to George Mason University: “Phylogenetic Diversity of Mycobionts and Photobionts in the Cyanolichen Genus Dictyonema, with Empasis on the Neotropics and the Galapagos Islands” (PI J. Lawrey; Co-PIs: R. Lücking, P. Gillevet). The biological material was collected as part of grant DEB-0715660 to The Field Museum: “Neotropical Epiphytic Microlichens—An Innovative Inventory of a Highly Diverse yet Little Known Group of Symbiotic Organisms” (PI R. Lücking). Naga Betrapally assisted with the SRA and BioProject submission of the dataset.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lücking, R., Lawrey, J.D., Gillevet, P.M. et al. Multiple ITS Haplotypes in the Genome of the Lichenized Basidiomycete Cora inversa (Hygrophoraceae): Fact or Artifact?. J Mol Evol 78, 148–162 (2014). https://doi.org/10.1007/s00239-013-9603-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-013-9603-y