Abstract
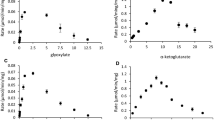
Procedures for the purification of an aldehyde dehydrogenase from extracts of the obligate methylotroph, Methylomonas methylovora are described. The purified enzyme is homogeneous as judged from polyacrylamide gel electrophoresis. In the presence of an artificial electron acceptor (phenazine methosulfate), the purified enzyme catalyzes the oxidation of straight chain aldehydes (C1-C10 tested), aromatic aldehydes (benzaldehyde, salicylaldehyde), glyoxylate, and glyceraldehyde. Biological electron acceptors such as NAD+, NADP+, FAD, FMN, pyridoxal phosphate, and cytochrome c cannot act as electron carriers. The activity of the enzyme is inhibited by sulfhydryl agents [p-chloromercuribenzoate, N-ethylmaleimide and 5,5-dithiobis (2-nitrobenzoic acid)], cuprous chloride, and ferrour nitrate. The molecular weight of the enzyme as estimated by gel filtration is approximately 45000 and the subunit size determined by sodium dodecyl sulfate-gel electrophoresis is approximately 23000. The purified enzyme is light brown and has an absorption peak at 410 nm. Reduction of enzyme with sodium dithionite or aldehyde substrate resulted in the appearance of peaks at 523 nm and 552 nm. These results suggest that the enzyme is a hemoprotein. There was no evidence that flavins were present as prosthetic group. The amino acid composition of the enzyme is also presented.
Similar content being viewed by others
Abbreviations
- PMS:
-
phenazine methosulfate
- DCPIP:
-
2,6-dichlorophenol indophenol
- DEAE:
-
diethylaminoethyl
References
Anthony, C., Zatman, L. J.: The microbial oxidation of methanol. Purification and properties of the alcohol dehydrogenase of Pseudomonas M27. Biochem. J. 104, 953–959 (1967a)
Anthony, C., Zatman, L. J.: The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas M27: A new oxidoreductase prosthetic group. Biochem. J. 104, 960–969 (1967b)
Basford, R. E., Huennekens, F. M.: Oxidation of thiol groups by 2,6-dichlorophenol-indophenol. J. Amer. Chem. Soc. 77, 3873–3877 (1955)
Boulton, C. A., Large, P. J.: Synthesis of certain assimilatory and dissimilatory enzymes during bacterial adaptation by growth on trimethylamine. J. Gen. Microbiol. 101, 151–156 (1977)
Colby, J., Zatman, L. J.: Hexose phosphate synthetase and tricarboxylic acid-cycle enzymes in bacterium 4B6, an obligate methylotroph. Biochem. J. 128, 1373–1376 (1972)
Colby, J., Zatman, L. J.: Trimethylamine metabolism in obligate and facultative methylotrophs. Biochem. J. 132, 101–112 (1973)
Foster, J. W., Davis, R. H.: A methane-dependent coccus, with notes on classification and nomenclature of obligate methane-utilizing bacteria. J. Bacteriol. 91, 1924–1931 (1966)
Hampton, D., Zatman, L. J.: The metabolism of tetramethylammonium chloride by bacterium 5H2. Biochem. Soc. Trans. 1, 667–668 (1973)
Harrington, A. A., Kallio, R. E.: Oxidation of methanol and formaldehyde by Pseudomonas methanica. Can. J. Microbiol. 6, 1–7 (1960)
Jakoby, W. B.: Aldehyde oxidation I. Dehydrogenase from Pseudomonas fluorescens. J. Biol. Chem. 232, 75–87 (1958)
Johnson, P. A., Quayle, J. R.: Microbial growth on C-1 compounds. Oxidation of methanol, formaldehyde and formate by methanolgrown Pseudomonas AM1. Biochem. J. 93, 281–290 (1964)
King, T. E., Cheldelin, V. H.: Oxidation of acetaldehyde by Acetobacter suboxydans. J. Biol. Chem. 220, 177–191 (1956)
Kung, H. F., Wagner, C.: Oxidation of C1 compounds by Pseudomonas sp. MS. Biochem. J. 116, 357–365 (1970)
Kuono, K., Oki, T., Nomura, H., Ozaki, A.: Isolation of new methanol-utilizing bacteria and its thiamine-requirement for growth. J. Gen. Appl. Microbiol. 19, 11–21 (1973)
Lowry, O. H., Rosenbrough, N. J., Farr, A. L., Randall, R. J.: Protein measurement with Folin phenol reagent. J. Biol. Chem. 193, 265–275 (1951)
Mahler, H. R., Mackler, B., Grew, D. E.: Studies on metalloflavoprotein. III. Aldehyde oxidase: a molybdoflavoprotein. J. Biol. Chem. 210, 465–480 (1954)
Matsubara, M., Sasaki, R. M.: High recovery of tryptophan from acid hydrolysis of proteins. Biochem. Biophys. Res. Commun. 35, 175–181 (1969)
Meagher, R. B., Ornston, L. N.: Relationship among enzymes of the β-keto-adipate pathway. I. Properties of cis, cis-muconate lactonizing enzyme and muconolacetone isomerase from Pseudomonas putida. Biochemistry 12, 3523–3530 (1973)
Mehta, R. J.: A novel inducible formaldehyde dehydrogenase of Pseudomonas sp. (RJ). Antonie van Leeuwenhoek, J. Microbiol. Serol. 41, 89–95 (1975)
Moore, S., Stein, W. H.: Chromatographic determination of amino acids by the use of automatic recording equipment. In: Methods in enzymology (S. P. Colowick and N. O. Kaplan, eds), Vol. 6, p. 819. New York: Academic Press, Inc. 1963
O'Connor, M. L., Hanson, R. S.: Enzyme regulation in Methylobacterium organophilum. J. Gen. Microbiol. 101, 327–332 (1977)
Patel, R. N., Bose, H. R., Mandy, W. J., Hoare, D. S.: Physiological studies of methane and methanol-oxidizing bacteria: Comparison of a primary alcohol dehydrogenase from Methylococcus capsulatus and Pseudomonas M27. J. Bacteriol. 107, 570–577 (1972)
Patel, R. N., Felix, A.: Microbial oxidation of methane and methanol: Crystallization and properties of methanol dehydrogenase from Methylosinus sporium. J. Bacteriol. 128, 413–424 (1976)
Raymond, S.: A convenient apparatus for vertical gel electrophoresis. Clin. Chem. 8, 455–470 (1962)
Rose, Z. B., Racker, E.: Formaldehyde dehydrogenase from Bakers yeast. J. Biol. Chem. 237, 3279–3284 (1962)
Spencer, R. L., Wold, F.: New convenient method for estimation of total cystine and cysteine in protein. Anal. Biochem. 32, 185–190 (1969)
Sperl, G. T., Forrest, H. S., Gibson, D. T.: Substrate specificity of the purified primary alcohol dehydrogenase from methanoloxidizing bacteria. J. Bacteriol. 118, 541–550 (1974)
Stirling, D. I., Dalton, H.: Purification and properties of an NADP+-linked formaldehyde dehydrogenase from Methylococcus capsulatus (Bath). J. Gen. Microbiol. 107, 19–29 (1978)
Strittmatter, P., Ball, E. G.: Formaldehyde dehydrogenase — a glutathione-dependent enzyme system. J. Biol. Chem. 213, 445–461 (1955)
Urushibara, T., Forrest, H. S., Hoare, D. S., Patel, R. N.: Pteridine produced by Methylococcus capsulatus. Isolation and identification of a neopterin 2′3′-phosphate. Biochem. J. 125, 14–146 (1971)
Weber, K., Osborn, M.: The reliability of molecular weight determination by dodecyl sulfate polyacrylamide gel electrophoresis. J. Biol. Chem. 244, 4406–4412 (1969)
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Patel, R.N., Hou, C.T. & Felix, A. Microbial oxidation of methane and methanol: Purification and properties of a heme-containing aldehyde dehydrogenase from Methylomonas methylovora . Arch. Microbiol. 122, 241–247 (1979). https://doi.org/10.1007/BF00411286
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00411286