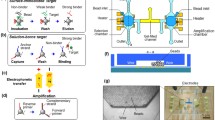
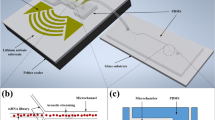
Abstract
Multi-target aptamer selection, known as multiplex systematic evolution of ligands by exponential enrichment (SELEX), is rapidly drawing interest because of its potential to enable high-speed, high-throughput aptamer selection. The parallelization of chemical processes by integrating microfluidic unit operations is a key strategy for developing a multiplex SELEX process. One of the potential problems with on-chip multiplexing chemical processes is cross-contamination. In order to avoid this, we propose a microfluidic network platform that uses pneumatic valves to allow the serial loading and incubation of aptamers with sol-gel entrapped target proteins. After target binding inside the sol-gels, the cross-contamination-free parallel elution of specifically bound aptamers is performed. The platform allows selective binding with five different targets immobilized in sol-gel spots. When eluting bound species, cross-contamination is avoided by sealing the adjacent elution chambers from each other using the pneumatic microvalves. Consequently, we demonstrate specific aptamer binding to the respective protein target and subsequent aptamer elution without any cross-contamination. This proof of concept opens the way to increased automation and microscale parallel processing of the SELEX methodology.
Similar content being viewed by others
References
Gold, L. et al. From oligonucleotide shapes to genomic SELEX: Novel biological-regulatory-loops. Proc. Natl. Acad. Sci. USA 94, 59–64 (1997).
Shi, H. et al. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl. Acad. Sci. USA 96, 10033–10038 (1999).
Tuerk, C. & MacDougal-Waugh, S. In vitro evolution of functional nucleic acids: high-affinity RNA ligands of HIV-1 proteins. Gene 137, 33–39 (1993).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Jo, M. et al. Development of single-stranded DNA aptamers for specific Bisphenol a detection. Oligonucleotides 21, 85–91 (2011).
Ahn, J.Y. et al. Aptamer microarray mediated capture and mass spectrometry identification of biomarker in serum samples. J. Proteome. Res. 9, 5568–5573 (2010).
Jenison, R.D. et al. High-resolution molecular discrimination by RNA. Science 263, 1425–1429 (1994).
Ahn, J.Y. et al. Selection of aptamers in SELEX process. Toxicol. Environ. Health Sci. 1, 1–7 (2009).
Fan, X. et al. Probing TBP interactions in transcription initiation and reinitiation with RNA aptamers that act in distinct modes. Proc. Natl. Acad. Sci. USA 101, 6934–6939 (2004).
Kwon, J. et al. High diagnostic accuracy of antigen microarray for sensitive detection of hepatitis C virus infection. Clin. Chem. 54, 424–428 (2008).
Hybarger, G. et al. A microfluidic SELEX prototype. Anal. Bioanal. Chem. 384, 191–198 (2006).
Lou, X. et al. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. USA 106, 2989–2994 (2009).
Ahn, J.Y. et al. A sol-gel-based microfluidics system enhances the efficiency of RNA aptamer selection. Oligonucleotides 21, 93–100 (2011)
Park, S.M. Selection and elution of aptamers using nanoporous sol-gel arrays with integrated microheaters. Lab. Chip 9, 1206–1212 (2009).
Thorsen, T. et al. Microfluidic large scale integration. Science 298, 580–584 (2002).
Hong, J.W. et al. Integrated nanoliter systems. Nat. Biotechnol. 21, 1179–1183 (2003).
Jeong, O. & Konishi, S. Pneumatic gas regulator with cascaded PDMS seal valves. Sens. Acts. A 143, 84–89 (2008).
Gill, I. & Ballesteros, A. Bioencapsulation within synthetic polymers (Part 1): sol-gel encapsulated biologicals. Trends in Biotechnology 18, 282–296 (2000).
Ahn, J.Y. et al. Sol-gel material optimization for aptamer biosensors. Mol. Cell. Toxicol. 4, 100–105 (2008).
Frenkel-Mullerad, H. & Avnir, D. Sol-gel materials as efficient enzyme protectors:? Preserving the activity of phosphatases under extreme pH conditions. J. Am. Chem. Soc. 127, 8077–8081 (2005).
Shi, H. et al. RNA aptamers directed to discrete functional sites on a single protein structural domain. Proc. Natl. Acad. Sci. USA 104, 3742–3746 (2007).
Sevilimedu, A. et al. TFIIB aptamers inhibit transcription by perturbing PIC formation at distinct stages. Nucleic Acids Res. 36, 3118–3127 (2008).
Mallik, P.K. et al. Commandeering a biological pathway using aptamer-derived molecular adaptors. Nucleic Acids Res. 38, e93 (2010).
Zhao, X. et al. An RNA aptamer that interferes with the DNA binding of the HSF transcription activator Nucleic Acids Res. 34, 3755–3761 (2006).
Kim, S. et al. Improved sensitivity and physical properties of sol-gel protein chips using large-scale material screening and selection. Anal. Chem. 78, 7392–7396 (2006).
Lee, S. et al. Chip-based detection of hepatitis C virus using RNA aptamers that specifically bind to HCV core antigen. Biochem. Biophys. Res. Commun. 358, 47–52 (2007).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Lee, S., Kang, J., Ren, S. et al. A cross-contamination-free SELEX platform for a multi-target selection strategy. BioChip J 7, 38–45 (2013). https://doi.org/10.1007/s13206-013-7106-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-013-7106-y