Abstract
Purpose
The enteric microbiome is known to play a major role in healthy gut homeostasis and several disease states. It may also contribute to both the intestinal recovery and complications that occur in patients with short bowel syndrome. The extent and nature of alterations to the gut microbiota following intestinal resection, however, are not well studied in a controlled setting. The purpose of this investigation is to characterize the effects of massive small bowel resection on the murine enteric microflora.
Methods
Wild-type C57BL6 mice, following a week of acclamation to a liquid rodent diet, underwent either 50 % proximal small bowel resection (SBR) or a sham operation. Mice were sacrificed, and enteric contents from the small bowel, cecum, and stool were harvested at 7 and 90 days post-operatively. DNA was isolated, and the V3–V5 regions of the 16s rRNA gene amplified and pyrosequenced on a Roche 454 platform. Sequences were clustered into operation taxonomic units and classified. Communities were then analyzed for diversity and phylogenic composition.
Results
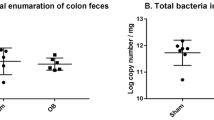
In the long-term group, the microbes inhabiting the ileum of mice undergoing SBR and sham operation differed significantly at the genus level (p < 0.001). Small bowel contents collected before and after SBR also differed significantly (p = 0.006). This was driven by an increase in Lactobacillus and decrease in Enterobacteriaceae species in mice undergoing SBR. No difference was seen in the long-term stool or in stool, cecal, or ileal contents in the short-term. No difference in microbial community diversity was found in any group.
Conclusion
Bowel resection induces long-term changes in the microbial community of the murine ileum, but not at more distal sites of the gastrointestinal tract. The increase in Lactobacillus encountered small bowel of resected mice correlates with limited previous studies. These changes may reflect an adaptive response of the microbiota to maximize energy extraction, but further studies are needed to establish the role played by this altered community.




Similar content being viewed by others
References
Squires, R.H., et al., Natural history of pediatric intestinal failure: initial report from the Pediatric Intestinal Failure Consortium. J Pediatr, 2012. 161(4): p. 723–8 e2.
Cole, C.R., et al., The rate of bloodstream infection is high in infants with short bowel syndrome: relationship with small bowel bacterial overgrowth, enteral feeding, and inflammatory and immune responses. J Pediatr, 2010. 156(6): p. 941–7, 947 e1.
Sentongo, T.A., R. Azzam, and J. Charrow, Vitamin B12 status, methylmalonic acidemia, and bacterial overgrowth in short bowel syndrome. J Pediatr Gastroenterol Nutr, 2009. 48(4): p. 495–7.
Kaufman, S.S., et al., Influence of bacterial overgrowth and intestinal inflammation on duration of parenteral nutrition in children with short bowel syndrome. J Pediatr, 1997. 131(3): p. 356–61.
Dicksved, J., et al., Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J, 2008. 2(7): p. 716–27.
Ridaura, V.K., et al., Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science, 2013. 341(6150): p. 1241214.
Kau, A.L., et al., Human nutrition, the gut microbiome and the immune system. Nature, 2011. 474(7351): p. 327–36.
Lupton, J.R., Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr, 2004. 134(2): p. 479–82.
Guarner, F. and J.R. Malagelada, Gut flora in health and disease. Lancet, 2003. 361(9356): p. 512–9.
Turnbaugh, P.J., et al., The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med, 2009. 1(6): p. 6ra14.
Helmrath, M.A., et al., Intestinal adaptation following massive small bowel resection in the mouse. J Am Coll Surg, 1996. 183(5): p. 441–9.
Lapthorne, S., et al., Gut microbial diversity is reduced and is associated with colonic inflammation in a piglet model of short bowel syndrome. Gut Microbes, 2013. 4(3): p. 212–21.
Human Microbiome Project, C., A framework for human microbiome research. Nature, 2012. 486(7402): p. 215–21.
Cole, J.R., et al., The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res, 2009. 37(Database issue): p. D141-5.
Oksanen J, B.G., Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MH, Wagner H, Vegan: community ecology package. 2013.
Venables, W.N., B.D. Ripley, and W.N. Venables, Modern applied statistics with S. 4th ed. Statistics and computing. 2002, New York: Springer. xi, 495 p.
White, J.R., N. Nagarajan, and M. Pop, Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol, 2009. 5(4): p. e1000352.
Devine, A.A., et al., Impact of ileocecal resection and concomitant antibiotics on the microbiome of the murine jejunum and colon. PLoS One, 2013. 8(8): p. e73140.
Bartosch, S., et al., Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol, 2004. 70(6): p. 3575–81.
Dethlefsen, L. and D.A. Relman, Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A, 2011. 108 Suppl 1: p. 4554–61.
Dibaise, J.K., R.J. Young, and J.A. Vanderhoof, Enteric microbial flora, bacterial overgrowth, and short-bowel syndrome. Clin Gastroenterol Hepatol, 2006. 4(1): p. 11–20.
Bouhnik, Y., et al., Bacterial populations contaminating the upper gut in patients with small intestinal bacterial overgrowth syndrome. Am J Gastroenterol, 1999. 94(5): p. 1327–31.
Cole, C.R. and T.R. Ziegler, Small bowel bacterial overgrowth: a negative factor in gut adaptation in pediatric SBS. Curr Gastroenterol Rep, 2007. 9(6): p. 456–62.
Galdeano, C.M. and G. Perdigon, The probiotic bacterium Lactobacillus casei induces activation of the gut mucosal immune system through innate immunity. Clin Vaccine Immunol, 2006. 13(2): p. 219–26.
Eizaguirre, I., et al., Probiotic supplementation reduces the risk of bacterial translocation in experimental short bowel syndrome. J Pediatr Surg, 2002. 37(5): p. 699–702.
Tolga Muftuoglu, M.A., et al., Effects of probiotics on experimental short-bowel syndrome. Am J Surg, 2011. 202(4): p. 461–8.
Hirayama, K., Ex-germfree mice harboring intestinal microbiota derived from other animal species as an experimental model for ecology and metabolism of intestinal bacteria. Exp Anim, 1999. 48(4): p. 219–27.
Liou, A.P., et al., Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med, 2013. 5(178): p. 178ra41.
Tantemsapya, N., et al., Body composition and metabolic changes associated with massive intestinal resection in mice. J Pediatr Surg, 2008. 43(1): p. 14–9.
Author information
Authors and Affiliations
Corresponding author
Additional information
Discussant
Dr. John C Alverdy (Chicago, IL): This is a very nice first foray into an important area that seeks to understand how the intestine adapts to massive resection by a group that has both a long standing interest and expertise in the field. The methods present were extremely well executed, and the data displayed along the standards of the field. As it is becoming increasingly apparent that our microbial partners with whom our intestines have co-evolved play a key role in all aspects of injury and recovery, this type of analysis is timely and will grow in scope. We can certainly expect this group with their established record of experience in this area to take a deeper dive into the changes in microbial community structure and function that accompany intestinal resection and its adaptation. While microbial diversity is one aspect of the structural changes in microbial communities, there are many changes whose significance remains to be defined. The increase in Lactobacillus, as a result of resection, might be interpreted as a mechanism by which the superorganism (host and microbe) coordinate protection against invading pathogens at a time of vulnerability. The changes in Enterococcus are also interesting, as many clinicians observe this as commensal to switch to a pathogen during complications from recovery. Like any good study, more questions are generated than answered. For example, when do the microbiota changes following small bowel resection become maladaptive to recovery? What influence do they have on nutrient absorption? Are microbial disturbances the cause of overgrowth of highly pathogenic strains? How do they contribute to the liver failure in short gut syndrome. As the technology advances, we will see more of such studies with increasingly greater levels of detail and molecular resolution. The major question for these groups is what are their future plans and what new techniques are on the horizon to answer the more difficult questions.
Closing Discussant
Dr. Sommovilla: Thank you for your thoughtful comments, Dr. Alverdy. As you mentioned, our study raises many questions. Many of the questions you bring up, especially those pertaining to causal relationships between short bowel syndrome, microbial communities, and nutrition, require a carefully controlled environment to address, so, as you mentioned, better tools are necessary to move beyond an observational study. Our laboratory plans to address these questions, at least partially, by working with germ-free mice colonized with collections of cultured bacteria from human donors. These tools will provide a number of advantages to our experimental designs. First, we can study more precisely the impact of bowel resection on a controlled human microbial community. Second, by addition and subtraction of specific microbes, we can establish more causal relationships between specific microbial perturbations, diets, and host phenotypes. We also plan to pair such experiments with more nuanced metabolic analyses in the host (body composition, fat absorption, and metabolomics) to further develop these relationships.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplemental Figure 1
Community comparisons at Phylum level. Principal coordinate analysis of short (A) and long-term (B) study arm comparing SBR vs sham communities of stool (left), cecum (center) and small bowel (right) contents at time of harvest. (C) Breakdown of predominant phyla of the small bowel contents of each group (GIF 184 kb)
Rights and permissions
About this article
Cite this article
Sommovilla, J., Zhou, Y., Sun, R.C. et al. Small Bowel Resection Induces Long-Term Changes in the Enteric Microbiota of Mice. J Gastrointest Surg 19, 56–64 (2015). https://doi.org/10.1007/s11605-014-2631-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11605-014-2631-0