Abstract
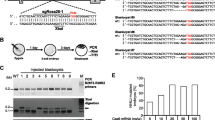
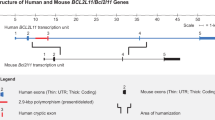
Transgenic mice expressing a tamoxifen-inducible Cre recombinase specifically in cardiomyocytes were generated in 2001 and are in widespread use, having been employed in >150 published studies. However, several groups recently have reported that tamoxifen administration to these mice can have off-target effects that include cardiac dysfunction, fibrosis, and death. For this reason, among others, we considered it important to better characterize the transgene (termed herein, CM-MCM) and its chromosomal location(s). Cytogenetic analysis positioned the CM-MCM transgene within the C band of chromosome 19, and more precise mapping, using genome walking and DNA sequencing, showed that transgene insertion is in the C1 region. Using the genome walking data, we have developed PCR assays that not only identify mice that carry the transgene, but also distinguish homozygous animals (CM-MCMTg/Tg) from hemizygous (CM-MCMTg/0), permitting the rapid assessment of transgene zygosity and, thereby, helping to minimize off-target tamoxifen-induced effects. Substantial rearrangement/duplication of transgene elements is present, and transgene integration was accompanied by the deletion of a 19,500 bp fragment of genomic DNA that contains the promoter, exon 1 and part of intron 1 of the APOBEC1 complementation factor (A1cf) gene, as well as several elements that are predicted to regulate chromosomal architecture. A1cf protein expression is ablated by the deletion and, therefore, homozygous mice are functionally A1cf knockout. The implications of this unexpected finding are discussed.



Similar content being viewed by others
References
Aldhous MC, Abu BS, Prescott NJ, Palla R, Soo K, Mansfield JC, Mathew CG, Satsangi J, Armour JA (2010) Measurement methods and accuracy in copy number variation: failure to replicate associations of beta-defensin copy number with Crohn’s disease. Hum Mol Genet 19:4930–4938
Althof N, Harkins S, Kemball CC, Flynn CT, Alirezaei M, Whitton JL (2014) In vivo ablation of type I interferon receptor from cardiomyocytes delays coxsackieviral clearance and accelerates myocardial disease. J Virol 88:5087–5099
Armour JA, Palla R, Zeeuwen PL, Den HM, Schalkwijk J, Hollox EJ (2007) Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res 35:e19
Bersell K, Choudhury S, Mollova M, Polizzotti BD, Ganapathy B, Walsh S, Wadugu B, Arab S, Kuhn B (2013) Moderate and high amounts of tamoxifen in αMHC-MerCreMer mice induce a DNA damage response, leading to heart failure and death. Dis Model Mech 6:1459–1469
Blanc V, Henderson JO, Newberry EP, Kennedy S, Luo J, Davidson NO (2005) Targeted deletion of the murine apobec-1 complementation factor (acf) gene results in embryonic lethality. Mol Cell Biol 25:7260–7269
Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, Mills RE, Kirby A, Lindgren AM, Rudiger SR, McLaughlan CJ, Bawden CS, Reid SJ, Faull RL, Snell RG, Hall IM, Shen Y, Ohsumi TK, Borowsky ML, Daly MJ, Lee C, Morton CC, MacDonald ME, Gusella JF, Talkowski ME (2012) Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet 44(390–7):S1
Dur S, Krause K, Pluntke N, Greeve J (2004) Gene structure and expression of the mouse APOBEC-1 complementation factor: multiple transcriptional initiation sites and a spliced variant with a premature stop translation codon. Biochim Biophys Acta 1680:11–23
Guescini M, Sisti D, Rocchi MB, Stocchi L, Stocchi V (2008) A new real-time PCR method to overcome significant quantitative inaccuracy due to slight amplification inhibition. BMC Bioinform 9:326
Hall ME, Smith G, Hall JE, Stec DE (2011) Systolic dysfunction in cardiac-specific ligand-inducible MerCreMer transgenic mice. Am J Physiol Heart Circ Physiol 301:H253–H260
Hougen K, Aronsen JM, Stokke MK, Enger U, Nygard S, Andersson KB, Christensen G, Sejersted OM, Sjaastad I (2010) Cre-loxP DNA recombination is possible with only minimal unspecific transcriptional changes and without cardiomyopathy in Tg(αMHC-MerCreMer) mice. Am J Physiol Heart Circ Physiol 299:H1671–H1678
Koitabashi N, Bedja D, Zaiman AL, Pinto YM, Zhang M, Gabrielson KL, Takimoto E, Kass DA (2009) Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circ Res 105:12–15
Lexow J, Poggioli T, Sarathchandra P, Santini MP, Rosenthal N (2013) Cardiac fibrosis in mice expressing an inducible myocardial-specific Cre driver. Dis Model Mech 6:1470–1476
Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH (1990) A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5:1743–1753
Mehta A, Kinter MT, Sherman NE, Driscoll DM (2000) Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol 20:1846–1854
Molkentin JD, Robbins J (2009) With great power comes great responsibility: using mouse genetics to study cardiac hypertrophy and failure. J Mol Cell Cardiol 46:130–136
Mouse genome informatics (2015) References associated with the Tg(Myh6-cre/Esr1*)1Jmk allele. http://www.informatics.jax.org/reference/allele/MGI:3050453
Ong CT, Corces VG (2014) CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15:234–246
Rochat B (2005) Role of cytochrome P450 activity in the fate of anticancer agents and in drug resistance: focus on tamoxifen, paclitaxel and imatinib metabolism. Clin Pharmacokinet 44:349–366
Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD (2001) Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res 89:20–25
Srivastava A, Philip VM, Greenstein I, Rowe LB, Barter M, Lutz C, Reinholdt LG (2014) Discovery of transgene insertion sites by high throughput sequencing of mate pair libraries. BMC Genom 15:367
Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA, Campbell PJ (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144:27–40
VanBrocklin MW, Robinson JP, Whitwam T, Guilbeault AR, Koeman J, Swiatek PJ, Vande Woude GF, Khoury JD, Holmen SL (2009) Met amplification and tumor progression in Cdkn2a-deficient melanocytes. Pigment Cell Melanoma Res 22:454–460
Acknowledgments
This work was supported by NIH grant R01 AI110621. This is manuscript number 29197 from the Scripps Research Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Harkins, S., Whitton, J.L. Chromosomal mapping of the αMHC-MerCreMer transgene in mice reveals a large genomic deletion. Transgenic Res 25, 639–648 (2016). https://doi.org/10.1007/s11248-016-9960-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-016-9960-6