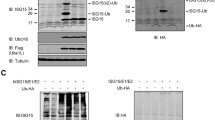
Abstract
Neuropathy target esterase (NTE) and NTE-related esterase (NRE) are endoplasmic reticulum (ER) membrane-anchored proteins belonging to the NTE protein family. NTE and NRE are degraded by macroautophagy and by the ubiquitin–proteasome pathway. However, the regulation of NTE and NRE by proteasome has not been well understood. Western blotting showed that the deletion of the regulatory region of NTE and NRE led to protein accumulation compared with that of the corresponding wild-type proteins. Further, deletion and site-directed mutagenesis experiments demonstrated that the destruction (D) box was required for the proteasomal degradation of NTE and NRE. However, unlike the deletion of the regulatory region, the deletion of the D box did not affect the subcellular localisation of NTE or NRE or disrupt the ER. Moreover, the deletion of the D box or the regulatory region of NTE has similar inhibitory effects on cell growth, which are greater than those produced by the full-length NTE. Here, for the first time, we show that the D box is involved in the regulation of NTE family proteins by the proteasome but not in their subcellular localisation. In addition, these results suggest that the NTE overexpression-mediated inhibition of cell growth is related to active protein levels but not to its ER disruption effect.





Similar content being viewed by others
References
Richardson RJ, Hein ND, Wijeyesakere SJ et al (2013) Neuropathy target esterase (NTE): overview and future. Chem Biol Interact 203:238–244
Lush MJ, Li Y, Read DJ et al (1998) Neuropathy target esterase and a homologous Drosophila neurodegeneration-associated mutant protein contain a novel domain conserved from bacteria to man. Biochem J 332:1–4
Winrow CJ, Hemming ML, Allen DM et al (2003) Loss of neuropathy target esterase in mice links organophosphate exposure to hyperactivity. Nat Genet 33:477–485
Wilson PA, Gardner SD, Lambie NM et al (2006) Characterization of the human patatin-like phospholipase family. J Lipid Res 47:1940–1949
Kienesberger PC, Oberer M, Lass A et al (2009) Mammalian patatin domain containing proteins: a family with diverse lipolytic activities involved in multiple biological functions. J Lipid Res 50:S63–S68
Li Y, Dinsdale D, Glynn P (2003) Protein domains, catalytic activity, and subcellular distribution of neuropathy target esterase in mammalian cells. J Biol Chem 278:8820–8825
Kienesberger PC, Lass A, Preiss-Landl K et al (2008) Identification of an insulin-regulated lysophospholipase with homology to neuropathy target esterase. J Biol Chem 283:5908–5917
Zaccheo O, Dinsdale D, Meacock PA et al (2004) Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J Biol Chem 279:24024–24033
Quistad GB, Barlow C, Winrow CJ et al (2003) Evidence that mouse brain neuropathy target esterase is a lysophospholipase. Proc Natl Acad Sci USA 100:7983–7987
Akassoglou K, Malester B, Xu J et al (2004) Brain-specific deletion of neuropathy target esterase/swiss cheese results in neurodegeneration. Proc Natl Acad Sci USA 101:5075–5080
Read DJ, Li Y, Chao MV et al (2009) Neuropathy target esterase is required for adult vertebrate axon maintenance. J Neurosci 29:11594–11600
Dutta S, Rieche F, Eckl N et al (2016) Glial expression of Swiss-cheese (SWS), the Drosophila orthologue of neuropathy target esterase, is required for neuronal ensheathment and function. Dis Model Mech 9:283–294
Rainier S, Bui M, Mark E et al (2008) Neuropathy target esterase gene mutations cause motor neuron disease. Am J Hum Genet 82:780–785
Synofzik M, Gonzalez MA, Lourenco CM et al (2014) PNPLA6 mutations cause Boucher–Neuhauser and gordon holmes syndromes as part of a broad neurodegenerative spectrum. Brain 137:69–77
Koh K, Kobayashi F, Miwa M et al (2015) Novel mutations in the PNPLA6 gene in Boucher-Neuhäuser syndrome. J Hum Genet 60:217–220
Topaloglu AK, Lomniczi A, Kretzschmar D et al (2014) Loss-of-function mutations in PNPLA6 encoding neuropathy target esteraseunderlie pubertal failure and neurological deficits in gordon holmes syndrome. J Clin Endocrinol Metab 99:E2067–E2075
Kmoch S, Majewski J, Ramamurthy V et al (2015) Mutations in PNPLA6 are linked to photoreceptor degeneration and various forms of childhood blindness. Nat Commun 6:5614
Hufnagel RB, Arno G, Hein ND et al (2015) Neuropathy target esterase impairments cause Oliver-McFarlane and laurence-moon syndromes. J Med Genet 52:85–94
Moser M, Li Y, Vaupel K et al (2004) Placental failure and impaired vasculogenesis result in embryonic lethality for neuropathy target esterase-deficient mice. Mol Cell Biol 24:1667–1679
Chen R, Chang PA, Long DX et al (2007) Down-regulation of neuropathy target esterase by protein kinase C activation with PMA stimulation. Mol Cell Biochem 302:179–185
Chen JX, Long DX, Hou WY et al (2010) Regulation of neuropathy target esterase by the cAMP/protein kinase a signal. Pharmacol Res 62:259–264
Chen JX, Wu YJ (2013) CREB is required for cAMP/PKA signals upregulating neuropathy target esterase expression. DNA Cell Biol 32:199–205
Chang PA, Wang ZX, Long DX et al (2012) Identification of two novel splicing variants of murine NTE-related esterase. Gene 497:164–171
Long DX, Chang PA, Liang YJ et al (2009) Degradation of neuropathy target esterase by the macroautophagic lysosomal pathway. Life Sci 84:89–96
Chang PA, Chen YY, Long DX et al (2012) Degradation of mouse NTE-related esterase by macroautophagy and the proteasome. Mol Biol Rep 39:7125–7131
Chang PA, Long DX, Wu YJ et al (2009) Identification and characterization of chicken neuropathy target esterase. Gene 435:45–52
Chang PA, Chen YY, Qin WZ et al (2011) The role of cell cycle-dependent neuropathy target esterase in cell proliferation. Mol Biol Rep 38:123–130
Long DX, Wang P, Sun YJ et al (2015) Neuropathy target esterase is degraded by the ubiquitin-proteasome pathway with ARA54 as the ubiquitin ligase. Biochemistry 254:7385–7392
Chen R, Chang PA, Long DX et al (2007) G protein β2 subunit interacts directly with neuropathy target esterase and regulates its activity. Int J Biochem Cell Biol 39:124–132
Glotzer M, Murray AW, Kirschner MW (1991) Cyclin is degraded by the ubiquitin pathway. Nature 349:132–138
Choi E, Dial JM, Jeong DE et al (2008) Unique D box and KEN box sequences limit ubiquitination of Acm1 and promotepseudosubstrate inhibition of the anaphase-promoting complex. J Biol Chem 283:23701–23710
Yamano H, TsurumiC GannonJ et al (1998) The role of the destruction box and its neighbouring lysine residues in cyclin B for anaphase ubiquitin-dependent proteolysis in fission yeast: defining the D-box receptor. EMBO J 17:5670–5678
Glynn P (2013) Neuronal phospholipid deacylation is essential for axonal and synaptic integrity. Biochim Biophys Acta 1831:633–1641
Chang PA, Liu CY, Chen R et al (2006) Effect of overexpression of neuropathy target esterase on mammalian cell proliferation. Cell Prolif 39:429–440
Acknowledgments
The authors would like to thank Dr. Paul Glynn for providing NTE constructs, and Enago (www.enago.cn) for the English language review.
Funding
This study was funded by the Natural Science Foundation Project of CQ CSTC (2014jcyjA10033, cstc2016jcyjA0572)), and by the Science and Technology Project from Chongqing Municipal Education Committee (KJ1400424).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Huang, FF., Chang, PA., Sun, LX. et al. The destruction box is involved in the degradation of the NTE family proteins by the proteasome. Mol Biol Rep 43, 1285–1292 (2016). https://doi.org/10.1007/s11033-016-4063-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-4063-2