Abstract
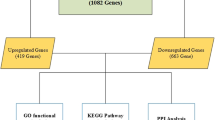
Lung cancer including non-small cell lung cancer (NSCLC) and small cell lung cancer is one of the most aggressive tumors with high incidence and low survival rate. The typical NSCLC patients account for 80–85 % of the total lung cancer patients. To systemically explore the molecular mechanisms of NSCLC, we performed a molecular network analysis between human and mouse to identify key genes (pathways) involved in the occurrence of NSCLC. We automatically extracted the human-to-mouse orthologous interactions using the GeneWays system by natural language processing and further constructed molecular (gene and its products) networks by mapping the human-to-mouse interactions to NSCLC-related mammalian phenotypes, followed by module analysis using ClusterONE of Cytoscape and pathway enrichment analysis using the database for annotation, visualization and integrated discovery (DAVID) successively. A total of 70 genes were proven to be related to the mammalian phenotypes of NSCLC, and seven genes (ATAD5, BECN1, CDKN2A, FNTB, E2F1, KRAS and PTEN) were found to have a bearing on more than one mammalian phenotype (MP) each. Four network clusters centered by four genes thyroglobulin (TG), neurofibromatosis type-1 (NF1 ), neurofibromatosis type 2 (NF2 ) and E2F transcription factor 1 (E2F1) were generated. Genes in the four network modules were enriched in eight KEGG pathways (p value < 0.05), including pathways in cancer, small cell lung cancer, cell cycle and p53 signaling pathway. Genes p53 and E2F1 may play important roles in NSCLC occurrence, and thus can be considered as therapeutic targets for NSCLC.





Similar content being viewed by others
References
Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63(1):11–30
Dempke W, Suto T, Reck M (2010) Targeted therapies for non-small cell lung cancer. Lung Cancer (Ams, Neth) 67(3):257
Strauss GM, Herndon J, Maddus MA et al (2004) Randomized clinical trial of adjuvant chemotherapy with paclitaxel and carboplatin following resection in stage IB non-small cell lung cancer (NSCLC): report of cancer and leukemia group B (CALGB) protocol 9633. J Clin Oncol (Meet Abstr) 22(Suppl 14):7019
Perez BA, Ghafoori AP, Lee C-L, Johnston SM, Li Y, Moroshek JG, Ma Y, Mukherjee S, Kim Y, Badea CT (2013) Assessing the radiation response of lung cancer with different gene mutations using genetically engineered mice. Front Oncol 3:72
Patrick O, Ferguson J, Shyu J, Current R, Rehage T, Tsai J, Christensen M, Tran HB, Chien SS-C, Shieh F (2013) Analytic performance studies and clinical reproducibility of a real-time PCR assay for the detection of epidermal growth factor receptor gene mutations in formalin-fixed paraffin-embedded tissue specimens of non-small cell lung cancer. BMC Cancer 13(1):210
Singh T, Katiyar SK (2013) Honokiol inhibits non-small cell lung cancer cell migration by targeting PGE2-mediated activation of β-catenin signaling. PLoS ONE 8(4):e60749
Yang X, Yao J, Luo Y, Han Y, Wang Z, Du L (2012) P38 MAP kinase mediates apoptosis after genipin treatment in non-small-cell lung cancer H1299 cells via a mitochondrial apoptotic cascade. J Pharmacol Sci 121(4):272–281
Chen H, Sharon E (2013) IGF-1R as an anti-cancer target—trials and tribulations. Chin J Cancer 32(5):242
Beer DG, Kardia SL, Huang C-C, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG (2002) Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 8(8):816–824
Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M (2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA 98(24):13790–13795
McDoniels-Silvers AL, Nimri CF, Stoner GD, Lubet RA, You M (2002) Differential gene expression in human lung adenocarcinomas and squamous cell carcinomas. Clin Cancer Res 8(4):1127–1138
Nacht M, Dracheva T, Gao Y, Fujii T, Chen Y, Player A, Akmaev V, Cook B, Dufault M, Zhang M (2001) Molecular characteristics of non-small cell lung cancer. Proc Natl Acad Sci USA 98(26):15203–15208
Rzhetsky A, Iossifov I, Loh JM, White KP (2006) Microparadigms: chains of collective reasoning in publications about molecular interactions. Proc Natl Acad Sci USA 103(13):4940–4945
Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI (2001) Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA 98(24):13784–13789
Rzhetsky A, Koike T, Kalachikov S, Gomez SM, Krauthammer M, Kaplan SH, Kra P, Russo JJ, Friedman C (2000) A knowledge model for analysis and simulation of regulatory networks. Bioinformatics 16(12):1120–1128
Rosell R, Monzo M, O’Brate A, Taron M (2002) Translational oncogenomics: toward rational therapeutic decision-making. Curr Opin Oncol 14(2):171–179
Rodríguez-Penagos C, Salgado H, Martínez-Flores I, Collado-Vides J (2007) Automatic reconstruction of a bacterial regulatory network using natural language processing. BMC Bioinform 8(1):293
Rzhetsky A, Seringhaus M, Gerstein M (2008) Seeking a new biology through text mining. Cell 134(1):9–13
Rzhetsky A, Iossifov I, Koike T, Krauthammer M, Kra P, Morris M, Yu H, Duboué PA, Weng W, Wilbur WJ (2004) GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J Biomed Inform 37(1):43–53
Liu K, Hogan WR, Crowley RS (2011) Natural language processing methods and systems for biomedical ontology learning. J Biomed Inform 44(1):163–179
Bult CJ, Eppig JT, Blake JA, Kadin JA, Richardson JE (2013) The mouse genome database: genotypes, phenotypes, and models of human disease. Nucleic Acids Res 41(D1):D885–D891
Smith CL, Goldsmith C-A, Eppig JT (2005) The mammalian phenotype ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol 6(1):R7
Rzhetsky A, Zheng T, Weinreb C (2006) Self-correcting maps of molecular pathways. PLoS ONE 1(1):e61
Iossifov I, Rodriguez-Esteban R, Mayzus I, Millen KJ, Rzhetsky A (2009) Looking at cerebellar malformations through text-mined interactomes of mice and humans. PLoS Comput Biol 5(11):e1000559
Rodriguez-Esteban R, Iossifov I, Rzhetsky A (2006) Imitating manual curation of text-mined facts in biomedicine. PLoS Comput Biol 2(9):e118
Smith CL, Eppig JT (2012) The mammalian phenotype ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm Genome 23(9–10):653–668
Nepusz T, Yu H, Paccanaro A (2012) Detecting overlapping protein complexes in protein–protein interaction networks. Nat Methods 9(5):471–472
da Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. doi:10.1038/nprot.2008.211
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT (2011) The mouse genome database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res 39(suppl 1):D842–D848
Nakagawa K, Conrad NK, Williams JP, Johnson BE, Kelley MJ (1995) Mechanism of inactivation of CDKN2 and MTS2 in non-small cell lung cancer and association with advanced stage. Oncogene 11(9):1843–1851
Giaccone G (1996) Oncogenes and antioncogenes in lung tumorigenesis. Chest 109(Suppl 5):130S–134S
Eymin B, Gazzeri S, Brambilla C, Brambilla E (2001) Distinct pattern of E2F1 expression in human lung tumours: E2F1 is upregulated in small cell lung carcinoma. Oncogene 20(14):1678–1687
Hann CL, Rudin CM (2008) Management of small-cell lung cancer: incremental changes but hope for the future. Cancer 22:1486–1492
Ma Y, Gu Y, Zhang Q, Han Y, Yu S, Lu Z, Chen J (2013) Targeted degradation of KRAS by an engineered ubiquitin ligase suppresses pancreatic cancer cell growth in vitro and in vivo. Mol Cancer Ther 12(3):286–294
Fiala O, Pesek M, Finek J, Benesova L, Belsanova B, Minarik M (2013) The dominant role of G12C over other KRAS mutation types in the negative prediction of efficacy of epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Cancer Genet 206(1):26–31
Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S (2008) Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 9(10):962–972
Verweij KJ, Zietsch BP, Medland SE, Gordon SD, Benyamin B, Nyholt DR, McEvoy BP, Sullivan PF, Heath AC, Madden PA (2010) A genome-wide association study of Cloninger’s temperament scales: implications for the evolutionary genetics of personality. Biol Psychol 85(2):306–317
Moalem J, Choi DX, Khanafshar E, Ruan DT, Woeber KA, Clark OH, Kebebew E (2011) Metastatic papillary thyroid cancer misdiagnosed as non-small cell lung cancer. Thyroid 21(3):319–323
Ray BK, Dhar S, Henry C, Rich A, Ray A (2013) Epigenetic regulation by Z-DNA silencer function controls cancer-associated ADAM-12 expression in breast cancer: cross-talk between MeCP2 and NF1 transcription factor family. Cancer Res 73(2):736–744
Haber D (1997) Tumour-suppressor genes: evolving definitions in the genomic age. Nat Genet 16(4):320–322
Doddrell RD, Dun X-P, Shivane A, Feltri ML, Wrabetz L, Wegner M, Sock E, Hanemann CO, Parkinson DB (2013) Loss of SOX10 function contributes to the phenotype of human Merlin-null schwannoma cells. Brain 136(2):549–563
Sekido Y, Pass HI, Bader S, Mew DJ, Christman MF, Gazdar AF, Minna JD (1995) Neurofibromatosis type 2 (NF2) gene is somatically mutated in mesothelioma but not in lung cancer. Cancer Res 55(6):1227–1231
Goel A, Chhabra R, Ahmad S, Prasad A, Parmar V, Ghosh B, Saini N (2012) DAMTC regulates cytoskeletal reorganization and cell motility in human lung adenocarcinoma cell line: an integrated proteomics and transcriptomics approach. Cell Death Dis 3(10):e402
Salgia R, Li J-L, Ewaniuk DS, Wang Y-B, Sattler M, Chen W-C, Richards W, Pisick E, Shapiro GI, Rollins BJ (1999) Expression of the focal adhesion protein paxillin in lung cancer and its relation to cell motility. Oncogene 18(1):67
Flowers S, Xu F, Moran E (2013) Cooperative activation of tissue-specific genes by pRB and E2F1. Cancer Res 73(7):2150–2158
Maqbool SB, Mehrotra S, Kolpakas A, Durden C, Zhang B, Zhong H, Calvi BR (2010) Dampened activity of E2F1-DP and Myb-MuvB transcription factors in Drosophila endocycling cells. J Cell Sci 123(23):4095–4106
Dyson N (1998) The regulation of E2F by pRB-family proteins. Genes Dev 12(15):2245–2262
Zhang L, Kavanagh BD, Thorburn AM, Camidge DR (2010) Preclinical and clinical estimates of the basal apoptotic rate of a cancer predict the amount of apoptosis induced by subsequent proapoptotic stimuli. Clin Cancer Res 16(17):4478–4489
Vandin F, Upfal E, Raphael BJ (2011) Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol 18(3):507–522
Matsuoka S, Yamaguchi M, Matsukage A (1994) D-type cyclin-binding regions of proliferating cell nuclear antigen. J Biol Chem 269(15):11030–11036
Bremner R, Zacksenhaus E (2010) Cyclins, Cdks, E2f, Skp2, and more at the first international RB tumor suppressor meeting. Cancer Res 70(15):6114–6118
Mitsudomi T, Steinberg S, Nau M, Carbone D, D’amico D, Bodner S, Oie H, Linnoila R, Mulshine J, Minna J (1992) p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7(1):171
Goldenberg-Furmanov M, Stein I, Pikarsky E, Rubin H, Kasem S, Wygoda M, Weinstein I, Reuveni H, Ben-Sasson SA (2004) Lyn is a target gene for prostate cancer: sequence-based inhibition induces regression of human tumor xenografts. Cancer Res 64(3):1058–1066
Xu Q, Leong J, Chua QY, Chi YT, Chow PKH, Pack DW, Wang CH (2013) Combined modality doxorubicin-based chemotherapy and chitosan-mediated p53 gene therapy using double-walled microspheres for treatment of human hepatocellular carcinoma. Biomaterials 34(21):5149–5162
Acknowledgments
We wish to express our warm thanks to Fenghe (Shanghai) Information Technology Co., Ltd. Their ideas and help gave a valuable added dimension to our research.
Author information
Authors and Affiliations
Corresponding author
Additional information
The Publisher and Editor retract this article in accordance with the recommendations of the Committee on Publication Ethics (COPE). After a thorough investigation we have strong reason to believe that the peer review process was compromised.
About this article
Cite this article
Li, J., Bi, L., Sun, Y. et al. RETRACTED ARTICLE: Text mining and network analysis of molecular interaction in non-small cell lung cancer by using natural language processing. Mol Biol Rep 41, 8071–8079 (2014). https://doi.org/10.1007/s11033-014-3705-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3705-5