Abstract
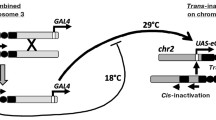
In salivary gland polytene chromosomes of Drosophila melanogaster, the regions of intercalary heterochromatin are characterized by late replication, under-replication, and genetic silencing. Using Gal4/UAS system, we induced transcription of sequences adjacent to transgene insertions in the band 11A6-9. This activation resulted in a loss of “silent” and appearance of “active” epigenetic marks, recruitment of RNA polymerase II, and formation of a puff. The activated region is now early replicating and shows increased level of DNA polytenization. Notably, all these changes are restricted to the area around the inserts, whereas the rest of the band remains inactive and late replicating. Although only a short area near the insertion site is transcribed, it results in an “open” chromatin conformation in a much broader region. We conclude that regions of intercalary heterochromatin do not form stand-alone units of late replication and under-replication. Every part of such regions can be activated and polytenized independently of other parts.






Similar content being viewed by others
References
Aggarwal BD, Calvi BR (2004) Chromatin regulates origin activity in Drosophila follicle cells. Nature 430:372–376
Ahmad K, Henikoff S (2001) Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104:839–847
Andreyeva EN, Kolesnikova TD, Belyaeva ES, Glaser RL, Zhimulev IF (2008) Local DNA underreplication correlates with accumulation of phosphorylated H2Av in the Drosophila melanogaster polytene chromosomes. Chromosome Res 16:851–862
Arcos-Teran L (1972) DNA replication and the nature of late replicating loci in the X-chromosome of Drosophila melanogaster. Chromosoma 37:233–296
Babenko VN, Makunin IV, Brusentsova IV, Belyaeva ES, Maksimov DA, Belyakin SN, Maroy P, Vasil'eva LA, Zhimulev IF (2010) Paucity and preferential suppression of transgenes in late replication domains of the D. melanogaster genome. BMC Genomics 11:318
Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, Hoskins RA, Spradling AC (2004) The BDGP gene disruption project: Single transposon insertions associated with 40 % of Drosophila genes. Genetics 167:761–781
Belyaeva ES, Demakov SA, Pokholkova GV, Alekseyenko AA, Kolesnikova TD, Zhimulev IF (2006) DNA underreplication in intercalary heterochromatin regions in polytene chromosomes of Drosophila melanogaster correlates with the formation of partial chromosomal aberrations and ectopic pairing. Chromosoma 115:355–366
Belyaeva ES, Andreyeva EN, Belyakin SN, Volkova EI, Zhimulev IF (2008) Intercalary heterochromatin in polytene chromosomes of Drosophila melanogaster. Chromosoma 117:411–418
Belyaeva ES, Goncharov FP, Demakova OV, Kolesnikova TD, Boldyreva LV, Semeshin VF, Zhimulev IF (2012) Late replication domains in polytene and non-polytene cells of Drosophila melanogaster. PLoS One 7(1):e30035
Belyakin SN, Christophides GK, Alekseyenko AA, Kriventseva EV, Belyaeva ES, Nanayev RA, Makunin IV, Kafatos FC, Zhimulev IF (2005) Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc Natl Acad Sci USA 102:8269–8274
Belyakin SN, Babenko VN, Maksimov DA, Shloma VV, Kvon EZ, Belyaeva ES, Zhimulev IF (2010) Gene density profile reveals the marking of late replicated domains in the Drosophila melanogaster genome. Chromosoma 119:589–600
Borowiec JA, Schildkraut CL (2011) Open sesame: Activating dormant replication origins in the mouse immunoglobulin heavy chain (Igh) locus. Curr Opin Cell Biol 23:284–292
Bridges C (1935) Salivary chromosome map with a key to the banding of the chromosomes of Drosophila melanogaster. J Hered 26:60–64
Cavalli G, Paro R (1998) The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93:505–518
Chambeyron S, Bickmore WA (2004) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18:1119–1130
Chintapalli VR, Wang J, Dow JAT (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nature Genet 39:715–720
Donaldson AD (2005) Shaping time: Chromatin structure and the DNA replication programme. Trends Genet 21:444–449
Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM (2011) Chromatin signatures of the Drosophila replication program. Genome Res 21:164–174
Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143:212–224
Gierman HJ, Indemans MHG, Koster J, Goetze S, Seppen J, Geerts D, van Drie R, Versteeg R (2007) Domain-wide regulation of gene expression in the human genome. Genome Res 17:1286–1295
Gilbert DM (2010) Cell fate transitions and the replication timing decision point. J Cell Biol 191:899–903
Graveley BR, Brooks AN, Carlson JW et al (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471:473–479
Guilbaud G, Rappailles A, Baker A, Chen CL, Arneodo A, Goldar A, d’Aubenton-Carafa Y, Thermes C, Audit B, Hyrien O (2011) Evidence for sequential and increasing activation of replication origins along replication timing gradients in the human genome. PLoS Comput Biol 7(12):e1002322
Hägele K, Kalisch WE (1974) Initial phases of DNA synthesis in Drosophila melanogaster. I. Differential participation in replication of the X chromosomes in males and females. Chromosoma 47:403–413
Hiratani I, Gilbert DM (2009) Replication timing as an epigenetic mark. Epigenetics 4:93–97
Holmquist GP (1987) Role of replication time in the control of tissue-specific gene expression. Am J Hum Genet 40:151–173
Kalisch WE, Hägele K (1976) Correspondence of banding patterns to 3H-thymidine labeling patterns in polytene chromosomes. Chromosoma 57:19–23
Kaufmann BP (1939) Distribution of induced breaks along the X-chromosome of Drosophila melanogaster. Proc Natl Acad Sci USA 25:571–577
Kolesnikova TD, Semeshin VF, Andreyeva EN, Zykov IA, Kokoza EB, Kalashnikova DA, Belyaeva ES, Zhimulev IF (2011) Induced decondensation of heterochromatin in Drosophila melanogaster polytene chromosomes under condition of ectopic expression of the Supressor of underreplication gene. Fly (Austin) 5:181–190
Madigan JP, Chotkowski HL, Glaser RL (2002) DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res 30:3698–3705
Makunin IV, Volkova EI, Belyaeva ES, Nabirochkina EN, Pirrotta V, Zhimulev IF (2002) The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics 160:1023–1034
MacAlpine DM, Bell SP (2005) A genomic view of eukaryotic DNA replication. Chromosome Res 13:309–326
MacAlpine DM, Rodríguez HK, Bell SP (2004) Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18:3094–3105
MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM (2010) Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20:201–211
Nordman J, Li S, Eng T, MacAlpine D, Orr-Weaver TL (2010) Developmental control of the DNA replication and transcription programs. Genome Res 21:175–181
Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL (2005) Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell 20:575–587
Petesch SJ, Lis JT (2008) Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134:74–84
Pope BD, Hiratani I, Gilbert DM (2010) Domain-wide regulation of DNA replication timing during mammalian development. Chromosome Res 18:127–136
Ranz JM, Díaz-Castillo C, Petersen R (2012) Conserved gene order at the nuclear periphery in Drosophila. Mol Biol Evol 29:13–16
Ryba T, Hiratani I, Sasaki T, Battaglia D, Kulik M, Zhang J, Dalton S, Gilbert DM (2011) Replication timing: A fingerprint for cell identity and pluripotency. PLoS Comput Biol 7(10):e1002225
Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schübeler D (2009) Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev 23:589–601
Schwaiger M, Schübeler D (2006) A question of timing: Emerging links between transcription and replication. Curr Opin Genet Dev 16:177–183
Sher N, Bell GW, Li S, Nordman J, Eng T, Eaton ML, Macalpine DM, Orr-Weaver TL (2012) Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res 22:64–75
Tumbar T, Sudlow G, Belmont AS (1999) Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol 145:1341–1354
White EJ, Emanuelsson O, Scalzo D, Royce T, Kosak S, Oakeley EJ, Weissman S, Gerstein M, Groudine M, Snyder M, Schübeler D (2004) DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc Natl Acad Sci USA 101:17771–17776
Zhimulev IF, Belyaeva ES, Makunin IV, Pirrotta V, Volkova EI, Alekseyenko AA, Andreyeva EN, Makarevich GF, Boldyreva LV, Nanayev RA, Demakova OV (2003) Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma 111:377–398
Acknowledgments
The authors are very grateful to Prof. K. McKim, Prof. E. Käs, and Prof. H. Saumweber for the antibodies; thank Bloomington Drosophila Stock Center and Prof. P.G. Georgiev for the flystocks; and thank Dr. A. Gorchakov for the discussion. This work was supported by the Program of Presidium of the Russian Academy of Sciences “Molecular and Cellular Biology” (grant no. 6.4) and by the grant of Russian Foundation of Basic Research 12-04-31128 (DAM).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Terry Orr-Weaver
Dmitry E. Koryakov and Galina V. Pokholkova contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Changes in chromatin make-up upon induced transcription of EY01976. Shown is a fragment of X chromosome (regions 10-11) with inactive insertion (a), insertion activated by hsp70-Gal4 at 18°С (b) and by da-Gal4 (c). Phase contrast (a-c) and overlay of phase contrast and immunostaining (d-h). Lines connect the same chromosome regions. Asterisk denotes a puff at 10EF. Bar, 5 μm (JPEG 59 kb)
Supplementary Fig. 2
Normalized RNA quantification around EY01976 insertion, shown as a log scale. qRT-PCR probes used and their positions on molecular map are indicated below the columns. Genotypes and induction conditions are color-coded. Error bars represent standard deviation (GIF 6 kb)
Supplementary Fig. 3
Degree of polytenization within the region 11A6-9 harboring EY01976 transgene, activated with da-Gal4 driver. x-axis shows genomic coordinates, y-axis shows the level of polytenization. The y-axis was scaled so that Actin42A gene polytenization level was equal 1. Red-shaded curves (red, orange, pink) correspond to the lines with inactive insertions or with Gal4-drivers. Blue-shaded lines (dark blue, light blue, purple) depict the polytenization profiles in stocks where transgenes are Gal4-activated. Arrows indicate insertion and PCR probes positions. Error bars represent standard deviation (GIF 32 kb)
Supplementary Fig. 4
Examples of typical immunostaining results with antibodies against RNA pol II, H3K9me2, and H3K9ac in the polytene sections 10-11. Fluorescent signal and phase contrast images are shown separately. The images with fluorescence clearly demonstrate the fact that prior to activation the region 11A6-9 (indicated by arrows) was not associated with these proteins. Bar, 5 μm (JPEG 99 kb)
Rights and permissions
About this article
Cite this article
Koryakov, D.E., Pokholkova, G.V., Maksimov, D.A. et al. Induced transcription results in local changes in chromatin structure, replication timing, and DNA polytenization in a site of intercalary heterochromatin. Chromosoma 121, 573–583 (2012). https://doi.org/10.1007/s00412-012-0382-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-012-0382-9