Abstract
The self-complementary subset \(\mathcal{T}_0 = \mathcal{X}_0 \)∪{AAA,TTT} with \(\mathcal{X}_0 \) = {AAC, AAT, ACC, ATC, ATT, CAG, CTC, CTG, GAA, GAC, GAG, GAT, GCC, GGC, GGT, GTA, GTC, GTT, TAC, TTC} of 22 trinucleotides has a preferential occurrence in the frame 0 (reading frame established by the ATG start trinucleotide) of protein (coding) genes of both prokaryotes and eukaryotes. The subsets \(\mathcal{T}_1 = \mathcal{X}_1 \)∪{CCC} and \(\mathcal{T}_2 = \mathcal{X}_2 \)∪{GGG} of 21 trinucleotides have a preferential occurrence in the shifted frames 1 and 2 respectively (frame 0 shifted by one and two nucleotides respectively in the 5′-3′ direction). \(\mathcal{T}_1 \) and \(\mathcal{T}_2 \) are complementary to each other. The subset \(\mathcal{T}_0 \) contains the subset \(\mathcal{X}_0 \) which has the rarity property (6 × 10−8) to be a complementary maximal circular code with two permutated maximal circular codes \(\mathcal{X}_1 \) and \(\mathcal{X}_2 \) in the frames 1 and 2 respectively. \(\mathcal{X}_0 \) is called a C3 code.
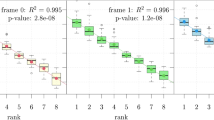
A quantitative study of these three subsets \(\mathcal{T}_0 ,\mathcal{T}_1 ,\mathcal{T}_2 \) in the three frames 0, 1, 2 of protein genes, and the 5′ and 3′ regions of eukaryotes, shows that their occurrence frequencies are constant functions of the trinucleotide positions in the sequences. The frequencies of \(\mathcal{T}_0 ,\mathcal{T}_1 ,\mathcal{T}_2 \) in the frame 0 of protein genes are 49, 28.5 and 22.5% respectively. In contrast, the frequencies of \(\mathcal{T}_0 ,\mathcal{T}_1 ,\mathcal{T}_2 \) in the 5′ and 3′ regions of eukaryotes, are independent of the frame. Indeed, the frequency of \(\mathcal{T}_0 \) in the three frames of 5′ (respectively 3′) regions is equal to 35.5% (respectively 38%) and is greater than the frequencies \(\mathcal{T}_1 \) and \(\mathcal{T}_2 \), both equal to 32.25% (respectively 31%) in the three frames.
Several frequency asymmetries unexpectedly observed (e.g. the frequency difference between \(\mathcal{T}_1 \) and \(\mathcal{T}_2 \) in the frame 0), are related to a new property of the subset \(\mathcal{T}_0 \) involving substitutions. An evolutionary analytical model at three parameters (p, q, t) based on an independent mixing of the 22 codons (trinucleotides in frame 0) of \(\mathcal{T}_0 \) with equiprobability (1/22) followed by t ≈ 4 substitutions per codon according to the proportions p ≈ 0.1; q ≈ 0.1 and r = 1 − p − q ≈ 0.8 in the three codon sites respectively, retrieves the frequencies of \(\mathcal{T}_0 ,\mathcal{T}_1 ,\mathcal{T}_2 \) observed in the three frames of protein genes and explains these asymmetries. Furthermore, the same model (0.1, 0.1, t) after t ≈ 22 substitutions per codon, retrieves the statistical properties observed in the three frames of the 5′ and 3′ regions. The complex behaviour of these analytical curves is totally unexpected and a priori difficult to imagine.
Similar content being viewed by others
References
Arquès, D. G. and C. J. Michel (1987). A purine-pyrimidine motif verifying an identical presence in almost all gene taxonomic groups. J. Theor. Biol. 128, 457–461.
Arquès, D. G. and C. J. Michel (1990). A model of DNA sequence evolution, Part 1: Statistical features and classification of gene populations, Part 2: Simulation model, Part 3: Return of the model to the reality. Bull. Math. Biol. 52, 741–772.
Arquès, D. G. and C. J. Michel (1992). A simulation of the genetic periodicities modulo 2 and 3 with processes of nucleotide insertions and deletions. J. Theor. Biol. 156, 113–127.
Arquès, D. G. and C. J. Michel (1993). Identification and simulation of new non-random statistical properties common to different eukaryotic gene subpopulations. Biochimie 75, 399–407.
Arquès, D. G. and C. J. Michel (1994). Analytical expression of the purine/pyrimidine autocorrelation function after and before random mutations. Math. Biosci. 123, 103–125.
Arquès, D. G. and C. J. Michel (1996). A complementary circular code in the protein coding genes. J. Theor. Biol. 182, 45–58.
Béal, M.-P. (1993). Codage Symbolique. Paris: Masson.
Béland, P. and T. F. H. Allen (1994). The origin and evolution of the genetic code. J. Theor. Biol. 170, 359–365.
Benne, R. (1989). RNA-editing in trypanosome mitochondria. Biochem. Biophys. Acta 1007, 131–139.
Benne, R., J. Van Den Burg, J. P. J. Brakenhoff, P. Sloof, J. H. Van Boom and M. C. Tromp (1986). Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46, 819–826.
Berstel, J. and D. Perrin (1985). Theory of Codes. New York: Academic Press.
Blaisdell, B. E. (1983). A prevalent persistent nonrandomness that distinguishes coding and non-coding eukaryotic nuclear DNA sequences. J. Mol. Evol. 19, 122–133.
Crick, F. H. C., S. Brenner, A. Klug and G. Pieczenik (1976). A speculation on the origin of protein synthesis. Origins of Life 7, 389–397.
Crick, F. H. C., J. S. Griffith and L. E. Orgel (1957). Codes without commas. Proc. Natl. Acad. Sci. 43, 416–421.
Dounce, A. L. (1952). Duplicating mechanism for peptide chain and nucleic acid synthesis. Enzymologia 15, 251–258.
Eigen, M. and P. Schuster (1978). The hypercycle. A principle of natural self-organization. Part C: The realistic hypercycle. Naturwissenschaften 65, 341–369.
Feagin, J. E. (1990). RNA editing in kinetoplastid mitochondria. J. Biol. Chem. 265, 19373–19376.
Feagin, J. E., J. M. Abraham and K. Stuart (1988). Extensive editing of the cytochrome c oxidase III transcript in trypanosoma brucei. Cell 53, 413–422.
Fickett, J. W. (1982). Recognition of protein coding regions in DNA sequences. Nucl. Acids Res. 10, 5303–5318.
Jukes, T. H. and V. Bhushan (1986). Silent nucleotide substitutions and G+C content of some mitochondrial and bacterial genes. J. Mol. Evol. 24, 39–44.
Konecny, J., M. Eckert, M. Schöniger and G. L. Hofacker (1993). Neutral adaptation of the genetic code to double-strand coding. J. Mol. Evol. 36, 407–416.
Konecny, J., M. Schöniger and G. L. Hofacker (1995). Complementary coding conforms to the primeval comma-less code. J. Theor. Biol. 173, 263–270.
Nirenberg, M. W. and J. H. Matthaei (1961). The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. 47, 1588–1602.
Shaw, J. M., J. E. Feagin, K. Stuart and L. Simpson (1988). Editing of kinetoplastid mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell 53, 401–411.
Shepherd, J. C. W. (1981). Method to determine the reading frame of a protein from the purine/pyrimidine genome sequence and its possible evolutionary justification. Proc. Natl. Acad. Sci. 78, 1596–1600.
Shulman, M. J., C. M. Steinberg and N. Westmoreland (1981). The coding function of nucleotide sequences can be discerned by statistical analysis. J. Theor. Biol. 88, 409–420.
Simpson, L. (1990). RNA editing—A novel genetic phenomenon? Science 250, 512–513.
Smith, T. F., M. S. Waterman and J. R. Sadler (1983). Statistical characterization of nucleic acid sequence functional domains. Nucl. Acids Res. 11, 2205–2220.
Staden, R. and A. D. McLachlan (1982). Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucl. Acids Res. 10, 141–156.
Stuart, K. (1991). RNA editing in mitochondrial mRNA of trypanosomatids. Trends Biochem. Sci. 16, 68–72.
Watson, J. D. and F. H. C. Crick (1953). A structure for deoxyribose nucleic acid. Nature 171, 737–738.
Zull, J. E. and S. K. Smith (1990). Is genetic code redundancy related to retention of structural information in both DNA strands? Trends Biochem. Sci. 15, 257–261.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Arqués, D.G., Fallot, JP. & Michel, C.J. An evolutionary analytical model of a complementary circular code simulating the protein coding genes, the 5′ and 3′ regions. Bull. Math. Biol. 60, 163–194 (1998). https://doi.org/10.1006/bulm.1997.0033
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1006/bulm.1997.0033