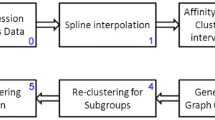
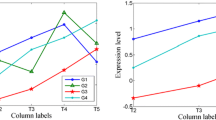
Abstract
Previous work for finding patterns only focuses on grouping objects under the same subset of dimensions. Thus, an important bio-interesting pattern, i.e. time-shifting, will be ignored during the analysis of time series gene expression data. In this paper, we propose a new definition of coherent cluster for time series gene expression data called ts-cluster. The proposed model allows (1) the expression profiles of genes in a cluster to be coherent on different subsets of dimensions, i.e. these genes follow a certain time-shifting relationship, and (2) relative expression magnitude is taken into consideration instead of absolute one, which can tolerate the negative impact induced by “noise”. This work is missed by previous research, which facilitates the study of regulatory relationships between genes. A novel algorithm is also presented and implemented to mine all the significant ts-clusters. Results experimented on both synthetic and real datasets show the ts-cluster algorithm is able to efficiently detect a significant amount of clusters missed by previous model, and these clusters are potentially of high biological significance.
Supported by the National Grand Fundamental Research 973 Program of China under Grant No. 2004BA721A05.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Hughes, T.R., Marton, M.J., et al.: Functional discovery via a compendium of expression profiles. Cell (2000)
Filkov, V., Skiena, S., Zhi, J.: Analysis techniques for microarray time-series data. In: 5th Annual International Conference on Computational Biology (2001)
Erdal, S., Ozturk, O., Armbruster, D., Ferhatosmanoglu, H., Ray, W.: A time series analysis of microarray data. In: 4th IEEE International Symposium on Bioinformatics and Bioengineering (May 2004)
Agrawal, R., Gehrke, J., Gunopulos, D., Raghavan, P.: Authomatic subspace clustering of high dimensional data for data mining applications. In: SIGMOD (1998)
Parsons, L., Haque, E., Liu, H.: Subspace clustering for high dimensional data: a review. In: SIGKDD (2004)
Cheng, Y., Church, G.M.: Biclustering of expression data. In: 8th International Conference on Intelligent Systems for Molecular Biology (2000)
Pei, J., Zhang, X., Cho, M., Wang, H., Yu, P.S.: Maple: A fast algorithm for maximal pattern-based clustering. In: ICDM 2003 Conf., Florida, pp. 259–266 (2003)
Wang, H., Yang, J., Wang, W., Yu, P.S.: Clustering by pattern similarity in large data sets. In: SIGMOD (2002)
Zhao, L., Zaki, M.J.: Tricluster: An effective algorithm for mining coherent clusters in 3d microarray data. In: ACM SIGMOD Conference (2005)
Yu, H., Luscombe, N.M., Qian, J., Gerstein, M.: Genomic analysis of gene expression relationships in transcriptional regulatory networks. Trends Genet. 19(8), 422–427 (2003)
Qiana, J., Dolled-Filharta, M., Lina, J., Yua, H., Gerstein, M.: Beyond synexpression relationships local clustering of time-shifted and inverted gene expression profiles identifies new, biologically relevant interactions. Journal of Molecular Biology 314(5), 1053–1066 (2001)
Spellman, P., Sherlock, G., et al.: Comprehensive identification of cell cycle-regulated genes of the yeast sacccharomyces cerevisiae by microarray hybridization. Molecular Biology of the Cell, 3273–3297 (1998)
Author information
Authors and Affiliations
Editor information
Rights and permissions
Copyright information
© 2007 Springer Berlin Heidelberg
About this paper
Cite this paper
Yin, Y., Zhao, Y., Zhang, B., Wang, G. (2007). Mining Time-Shifting Co-regulation Patterns from Gene Expression Data . In: Dong, G., Lin, X., Wang, W., Yang, Y., Yu, J.X. (eds) Advances in Data and Web Management. APWeb WAIM 2007 2007. Lecture Notes in Computer Science, vol 4505. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-72524-4_10
Download citation
DOI: https://doi.org/10.1007/978-3-540-72524-4_10
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-72483-4
Online ISBN: 978-3-540-72524-4
eBook Packages: Computer ScienceComputer Science (R0)