Abstract
Paleoecologists and paleogeographers still make use of binary coefficients in multivariate analysis decades after being introduced to the geosciences. Among the main groups, similarity, matching and association, selecting a particular coefficient remains a confusing and sometimes empirical process. Coefficients within groups tend to correlate highly when applied to datasets. With increasing interest in a probabilistic approach to grouping taxa or faunal lists, the Raup-Crick measure of association is closely related in purpose and empirically to coefficients of association and works well in cluster analysis and ordination. A reasonable strategy is to compare dendrograms and ordinations calculated with several coefficients, care being taken to select coefficients with different performance characteristics. Above all, the practitioner should understand the purpose of each coefficient.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
1 Introduction
Founding of the International Association for Mathematical Geology resulted in part from the increased use of quantitative methods in the geosciences and simultaneously with developments in computer hardware and availability. This is no less true for paleontology and paleoecology, fields of endeavor characterized by observing, describing, and synthesizing. With the 1960s and 70s came the development of large databases of fossil occurrences from which researchers could formally infer periods of rapid evolution and episodes of major extinction. Patterns of extinction through time could be simulated with random number generators. Paleoecologists studied whether fossil communities persisted through time and the structure of these communities.
This was a period of synthesis. The Treatise on Invertebrate Paleontology (Moore et al. 1953–2015) provided a need for stable taxonomies, a confidence that such a classification could be created, and the motivation for explaining trends in evolution.
Multivariate statistical methods developed in other fields became tools for reducing large datasets to manageable size while providing some degree of objectivity in the analysis. Cluster analysis, multidimensional scaling, factor analysis, and related eigenvector methods became familiar tools to the quantitatively-inclined geologist.
All methods required a measure of similarity, correlation, distance, dissimilarity, or association expressed as a coefficient. Eigenvector-based methods such as factor analysis and principal components analysis (PCA) by definition utilize covariance or correlation among variables (R-mode) and implicitly Euclidean distance in displays of sample coordinates (Q-mode). In contrast, cluster analysis, multidimensional scaling, and principal coordinates analysis (Gower 1971) allow use of a wide range of coefficients, but also require the user to decide which coefficient to use.
Multivariate statistical methods introduced to paleontologists in the 1960s and 70s continue to be used for studying the distribution of fossils in space and through time. With the existence of large databases of fossil occurrences, binary coefficients remain important for comparing collections made over many decades by many individuals.
It still remains to the practitioner to select one coefficient out of the many proposed over the course of more than a century now. Given no clear criterion, some elect to use several coefficients to see whether they affect results.
There is certainly a rich and extensive literature related to the purpose and performance of coefficients, both within the paleoecology literature and in the scientific and engineering literature at large as new applications are found for these coefficients. Surveys of existing measures range in approach, from considerations of the conceptual basis for each, to how well they satisfy the purpose, how they behave relative to each other, to how well they behave relative to a goal set by the author, whether or not they achieve a clear criterion such as satisfying metric properties (Gower and Legendre 1986), which seem to give similar results with each other or are correlated; and above all, whether coefficients include mutual absences. That last criterion might appear to be a small detail compared to the other comparisons but it introduces a fundamental question about the role that chance plays in the distribution of fossils in the collection under study.
This chapter reviews the criteria and arguments used in the past four decades in comparing binary coefficients. In this chapter, I will first group coefficients into three families based on shared formulations and behavior; discuss how such factors as abundance of taxa or poor sampling can affect coefficients; consider metric properties of coefficients; look at probability-based coefficients; apply several coefficients to paleoecological data; and sum up where we are today compared to four decades ago.
I will introduce coefficients as I go along, using what has become standard notation for binary coefficients. Assume we have sampled taxa from N locations. Then for a given pair of taxa:
-
a = number of co-occurrences
-
b = number of locations where taxon 1 occurs and taxon 2 does not
-
c = number of locations where taxon 2 occurs and taxon 1 does not
-
d = number of locations where neither taxon is observed
-
\( N = a + b + c + d \).
2 Empirical Comparisons and a Taxonomy
As the use of cluster analysis has expanded beyond the biological and geological sciences, papers have appeared in the literature that try to get a handle on the multitude of coefficients by comparing the way they behave relative to each other or to a criterion based on an application. Although outside the field of paleoecology, these publications often cast a wide net in gathering coefficients and present surveys purely empirical in nature. In the general area of pattern recognition, Choi et al. (2009) compute correlation coefficients among 76 binary coefficients for several types of random or structured datasets, observing that pairwise correlations between coefficients can be very high, depending in part on the pattern and number of presences.
In a companion paper, Choi et al. (2010) created random binary datasets, computed values for each coefficient, averaged the trials to create a dendrogram of the 76 coefficients. They identify eleven clusters, some with only a single coefficient, several with two to six members, and two large clusters with over twenty members. The second largest includes such frequently-used coefficients as the Jaccard, Otsuka, Dice, and the Bray and Curtis, where:
That these coefficients are correlated highly in an absolute sense should come as no surprise given the algebraic relationships between several. For example:
converting a dissimilarity coefficient (Bray and Curtis) into a coefficient expressing similarity. The difference between the Dice and Jaccard coefficients is in weighting the mutual occurrences. Remember that many coefficients were defined as measures of similarity, dissimilarity, or association rather than as input to clustering and ordination routines. Their creators had specific reasons for selecting and weighting the terms—a, b, c, or d—in the context of a study and according to some research goal. In many cases they might have been fully aware that their coefficient was similar to one in the literature, but their coefficient measured what they wanted to measure.
The largest group of coefficients includes a subset with among others the Simple Matching (called the Sokol and Michener in their paper), Rogers and Tanimoto, and Hamann coefficients, where:
Notice that these coefficient include the term d for mutual absence. The Rogers and Tanimoto coefficient is the same as the Simple Matching but for increased weighting for mismatches in the denominator and the Hamann can be expressed in terms of the Simple Matching by substituting N − (a + d) for (b + c).
A third, small group includes three similar coefficients, two derived from the familiar χ2 statistic, including the Phi coefficient:
These coefficients express correlation; in fact CPhi is the correlation coefficient for binary data and can be calculated in the same way as a correlation coefficient for non-binary data.
Related to these coefficients is a large cluster characterized by a numerator containing the term (ad – bc) or ad or (a + d). Examples are the Yule’s Q (or simply Yule), Ochiai 2, and Gower:
Similar to the matching coefficients, these and the Phi express agreement between two entities based on mutual presence and absence, but adjusted for relative abundance of the entities, analogous to the centering and scaling in calculating the correlation coefficient and Phi.
These four groups account for most of the binary coefficients one is likely to encounter in the geosciences, including ones discussed below. If we lump the last two clusters, a simple taxonomy of coefficients has as groups:
-
1.
Similarity coefficients, computed by the number of mutual occurrences, scaled by the total number of features occurring in one or the other entities. In paleoecology, entities can be taxa and features can be locations. Some coefficients can express similarity by calculating b + c rather than a, but the coefficient can be converted to similarity by subtracting from 1.
-
2.
Matching coefficients, generally expressing agreement by including both mutual occurrences and mutual absences d. Generally these are scaled to yield values between 0 and 1, but not always. For instance, the City Block or Hamming coefficient, b + c is not scaled. These are sometimes called distances (e.g. Hohn 1976) but as they are not Euclidean, this is not strictly correct. Although expressing disagreement, coefficients such as the City Block can be converted to matching coefficients.
-
3.
Coefficients of association, expressing how entities tend to vary together, adjusted for abundance or rarity.
This taxonomy agrees with that in Hohn (1976) except I am using a more rigorous definition of a distance coefficient by not including the City Block metric in that group. However, √(b + c) is a distance.
Even when two coefficients are not mathematically equivalent, they can be related monotonically (Gower and Legendre 1986) and give virtually the same results when used in cluster analysis or nonmetric multidimensional scaling. In lieu of selecting a single best coefficient, many researchers perform multiple cluster analyses or ordinations to observe whether results change with choice of coefficient. In such an exercise, one wants to make sure to select coefficients with different properties or behaviors.
3 Effects of Rare and Endemic Taxa
In an empirical study of eight similarity coefficients, Jackson et al. (1989) used a dataset comprised of 25 species of fish observed in 52 lakes in south-central Ontario, Canada. One feature that distinguishes this dataset is that species range from very common to rare, from as many as 47 lakes to as few as 2. The eight coefficients are the Jaccard, Dice, Simple Matching, Rogers and Tanimoto, Otsuka (“Ochiai” in their paper), Phi, Yule, and the Russell and Rao:
Unsurprisingly, the Jaccard and Dice gave nearly identical results in a cluster analysis. The same held for the Simple Matching and Rogers and Tanimoto coefficients. Results for the Otsuka were close to the Jaccard and Dice. The dendrogram for Russell and Rao coefficient shows almost no clusters although the general ordering of the species was very similar to the Jaccard, Dice, Simple Matching, Rogers and Tanimoto, and Otsuka.
They also performed principal coordinates analysis for each of the eight coefficients. They observed that the order of species on the first axis correlated highly with the number of lakes in which each occurred for all but the Otsuka, Phi, and Yule coefficients. Some of the correlations are very high, over 0.99 for the Simple Matching and Rogers and Tanimoto. In other words, the first axis corresponded to the frequency of each species, a general “size” factor in their words. Species abundance correlated poorly with the two major principal coordinates axes for the two coefficients of association, the Phi and Yule. The Otsuka showed some effect of species frequency. Nonmetric Multidimensional Scaling gave similar results. The order of species in dendrograms from cluster analysis also showed this frequency effect for the similarity coefficients; not so for the two coefficients of association.
They concluded that similarity coefficients—what they term co-occurrence coefficients—are heavily influenced by frequency, whereas the implicit centering that takes place in calculating the Phi and Yule mitigate this effect. They also conclude that the Otsuka formulation does a centering that partially eliminates the frequency effect.
4 Adjusting for Poor Sampling
In the context of Q-mode analysis—that is, the comparison of samples rather than the R-mode comparison of taxa—Alroy (2015a, b) looks at the effect of uneven sampling and consequent uneven sample size on four binary coefficients: the Forbes, a modified Forbes coefficient, Simpson’s coefficient, and the Dice, where the Forbes coefficient is:
and the Simpson:
Alroy modifies the Forbes coefficient in two ways. First, he argues against including mutual absences and therefore substitutes n for N where n = a + b + c. Secondly, he adds constants to correct for an upward bias in the coefficient:
Although there is no theoretical basis for these constants, the resulting coefficient does accomplish what he sets out to do. In several analyses of real and simulated datasets, he shows that both versions of the Forbes coefficient and the Simpson far outperform the Dice. This is consistent with results obtained by Jackson et al. (1989) in which coefficients such as the Dice are influenced very much by species frequency in R-mode analysis.
Alroy clearly favors the modified Forbes over Simpson’s coefficient. However results for both in cluster analysis and principal components analysis are very similar and would probably lead to the same conclusions based on the relative positions of samples on dendrograms and principal coordinates axes. This is no surprise given that the Simpson was formulated to account for uneven sample size.
Although Alroy dismisses probabilistic coefficients and coefficients of association in part for including mutual absences, it would be interesting to compare them with the two Forbes and the Simpson coefficients with his datasets.
These papers address the problem of working with datasets of mixed, perhaps unknown sampling regimen. The difference between otherwise identical faunal lists might be the time or skill in observation. This is perhaps less of an issue when a dataset comes from a single sampling campaign, but in these days of large databases compiled from many studies this is a problem to be taken seriously. Alroy’s results argue for careful selection of a coefficient and suggest that analysis with multiple coefficients might be beneficial if sampling issues are suspected.
Alroy points out that the Forbes coefficient has fallen out of use over time. However, since the publication of his papers, Halliday et al. (2017) used his modified form of the Forbes coefficient in cluster analysis of Late Cretaceous vertebrates across India. Although the papers by Alroy and by Halliday et al. describe ordination and cluster analysis of localities, the same problem of uneven sampling exists in analysis of taxa and their arguments and findings should have application in R-mode analysis as well.
5 Metric? Euclidean?
Some attention has been paid in the past with the question whether a dissimilarity coefficient is metric, Euclidean, or neither. A coefficient is metric if for every triplet (i, j, k) the following inequality holds:
On the face of it, methods such as principal coordinates analysis require a dissimilarity that is Euclidean. In actuality, Gower and Legendre (1986) and others have observed that departures from strict Euclidean geometry for many coefficients are generally small. Adding a constant to a distance can sometimes take care of this problem. It sometimes works to use the square root of the distance. They include a table showing that many familiar similarity coefficients, C, are metric but not Euclidean if converted to a dissimilarity coefficient 1 – C and even more are metric and many Euclidean if √(1 – C) is calculated. They consider most of the binary coefficients listed above with the notable exception of Yule’s coefficient.
Zhang and Srihari (2003) discuss the properties and behavior of similarity, matching, and coefficients of association, including metric properties, equivalent measures of similarity and dissimilarity, discriminatory capability of the coefficients, and the effect of weighting mutual absences. Like many authors they prefer metric coefficients. A large proportion of papers in the geosciences utilize nonmetric multidimensional scaling or cluster analysis with no requirement for the coefficient to be Euclidean or even metric. Reasons for selecting a method for multivariate analysis no doubt vary among authors, ranging from convenience or familiarity, available methods in a statistical package, to wanting to avoid the stronger requirements of eigenvector-based methods. However as Gower and Legendre (1986) point out, proportionally small deviations from geometric assumptions of an eigenvector method affects the results very little.
6 From Expected Values to Null Association
We can look at the diversity of coefficients along a spectrum from similarity coefficients at one end to coefficients of association at the other. In comparing faunal lists, for instance, similarity coefficients count the number of species in common between two locations normalized by the number of species found in one or the other. In other words, they can be said to measure overlap in faunal lists in a Q-mode analysis or geographic overlap of two taxa in an R-mode analysis.
Midway along the spectrum are coefficients that compare an observed value with the expected value. As described by Alroy (2015a), the chance of a species appearing in the faunal list at one site is (a + b)/N, the chance at a second site (a + c)/N, and the chance of being found in both is [(a + b) (a + c)]/N2. Therefore, the number of species expected to be found in both is [(a + b) (a + c)]/N and the ratio of the observed number a to the expected number is aN/[(a + b)(a + c)].
Hohn (1976), Raup and Crick (1979) and others have argued that cluster analysis or ordination should consider whether observed overlaps in faunal lists in paleogeographic studies or occurrence of taxa in paleoecological studies represent anything more than a random distribution of taxa through space. Of course there is no denying that species respond to environmental and geographic variables, but the question is how to separate similar distributions that arose by chance from those that represent nonrandom processes.
Within a biological context, Hubálek (1982) surveyed forty-three coefficients, eliminated about half based on algebraic equivalence, mere difference in scale, or failure to meet several criteria, and compared the rest through product-moment correlation and cluster analyses. Although one of these criteria is monotonicity with √(χ2), Hubálek stops short of recommending a coefficient such as Phi that is related directly to a test of significance in association.
In contrast, I proposed (Hohn 1976) that we should pay more attention to the Phi coefficient. Raup and Crick (1979) derive the formula for exact probabilities equal to Fisher’s Exact Test for independence in 2 by 2 tables, an alternative to the usual χ2 test. They modified what is essentially a Phi coefficient in comparing faunal lists by using a Monte Carlo method to weight taxa according by abundance. The result is a coefficient that like Phi and similar coefficients includes mutual absences, but represents a further refinement by taking relative abundance of taxa into account.
Winrow and Sutton (2014) calculated five coefficients—Raup-Crick, Simpson, Jaccard, Dice, and Otsuka (Ochiai)—in a paleogeographic study of lingulate brachiopods during the Early Paleozoic. Unable to determine a single best coefficient, they opted to calculate and compare several. Unsurprisingly, the Jaccard, Dice, and Otsuka gave very similar results. Raup-Crick and Simpson coefficients showed different patterns among pairs of faunal lists representing different paleocontinents. They do not explain why coefficients would give different results other than attributing several anomalously-high values of the Simpson to small sample sizes.
Zhang and Srihari (2003) survey binary dissimilarity coefficients in the context of character recognition; some of their results are instructive. In their look at nine familiar coefficients they define relative discriminatory power in terms of entropy, itself proportional to the variance of dissimilarities in multivariate space. They consider coefficients with a wide range of values to have potentially greater discriminatory power, finding that the Russell and Rao coefficient had the poorest discriminatory power and the Jaccard and related coefficients moderate power. Highest discriminatory power was shared by the correlation coefficient, Yule and Rogers and Tanimoto. In the study by Winrow and Sutton, the similarity coefficients had a narrow range of values compared with the Raup-Crick and Simpson.
7 Illustrative Example
Both R-mode and Q-mode analysis were performed on presence-absence data collected from five outcrops of the Middle Devonian Hamilton Group in New York State, although only ordinations of taxa will be shown here for reasons of space. Lithology of the interval sampled included thin limestones, mudstones, silty mudstones, and calcareous siltstones. The data matrix comprises 43 samples and 32 taxa identified to species when possible (Hohn 1975).
Cluster analysis, principal components analysis, and principal coordinates analysis were carried out; results of principal coordinates analysis best illustrate similarities and differences among the coefficients used. The statistical package PAST (Hammer et al. 2001) offers a wide range of multivariate methods and coefficients including similarity, matching, and association. I looked at results for the Phi (Correlation Coefficient in PAST) and Raup-Crick coefficients to observe their near-equivalence; the Jaccard as representative of similarity coefficients; Simpson’s coefficient as an unusual asymmetric coefficient used with some frequency; and to represent matching coefficients, the Hamming normalized to lie between 0 and 1:
In signal processing and information theory, Richard Hamming is known for the Hamming distance and Hamming window in addition to other contributions. Note the simple relationship between the normalized Hamming and Simple Matching coefficients:
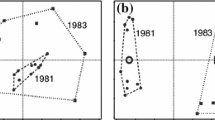
Looking at plots of the first two principal coordinate axes (Figs. 8.1, 8.2, 8.3, 8.4 and 8.5), one might be struck by the how similar they appear. However, most of us would probably consider the results from the Hamming (Simple Matching) coefficient in Fig. 8.1 difficult to interpret. The Jaccard is a great improvement (Fig. 8.2) as indeed is Simpson’s coefficient (Fig. 8.3). The Phi coefficients of association and Raup-Crick probabilistic measure give almost identical results with each other (Figs. 8.4 and 8.5).
The biggest differences among the five plots are positions of the most abundant taxa such as the brachiopod Tropidoleptus and bivalve Paleoneilo. They occur in a large proportion of samples (Table 8.1) and provide little discriminatory power among assemblages. Relatively abundant taxa score highly in an absolute sense on the second principal coordinate axis (vertical axis) for the Hamming and Jaccard coefficients, less so for the Raup-Crick and Phi. There is a clear correlation between principal coordinate scores on this axis with taxon count for the Hamming and Jaccard coefficients (Fig. 8.6). This observation agrees with the findings of Jackson et al. (1989).
Based on percent of variance explained by the first three principal coordinate axes (Table 8.2), the Hamming coefficient would appear to perform best. Similar results were obtained from nonmetric multidimensional scaling of each coefficient matrix (Table 8.3). But we already know that a portion of the variance correlates with taxon abundance. This observation suggests that selecting a coefficient based by variance explained has limited value if the coefficient measures the wrong thing.
Q-mode analyses showed similar correlation of abundance with principal coordinate scores calculated from Hemming and Jaccard coefficients. The relationship is not as strong because no sample contained more than 26% of the taxa, whereas Tropidoleptus in the R-mode analysis occurred in 84% of samples.
Note that the Raup-Crick procedure does not yield a binary coefficient in the sense of all the others, but rather accomplishes through Monte Carlo sampling, a similar measure as the correlation coefficient. Practitioners use the Raup-Crick measure in the same way as any of the other binary coefficients for cluster analysis and ordination. However there is no guarantee that it has strictly metric properties, and indeed, principal coordinates analysis with the Raup-Crick statistic yielded a large proportion of negative eigenvalues.
8 Discussion and Conclusions
Studies published over the past decade give a taste of the application of binary coefficients of all types.
Brayard et al. (2007) used distances 1 – SDice in Q-mode cluster analysis and ordination of Early Triassic ammonoid faunas, citing the double weight given to mutual presences, thus downweighting the influence of unique species occurrences and not giving any weight to mutual absences. They used the square root of the dissimilarity matrix so that the resulting distances would be metric and Euclidean (Gower and Legendre 1986).
In studies of faunal lists of bivalves from around the globe, Schmachtenberg (2008) compared four coefficients: Jaccard, Simpson, Raup-Crick, and a measure of endemism. He did not do any cluster analyses or ordinations, but rather regressed value of each coefficient on geographic distance. The Simpson, Raup-Crick, and natural log of the Jaccard coefficient performed almost equally well.
Huang et al. (2012) considered the performance of five coefficients—Jaccard, Dice, Cosine, Yule’s Y, and Raup-Crick—in cluster analysis and nonmetric multidimensional scaling of Silurian brachiopod assemblages representing time after the Late Ordovician extinction events. They preferred the Raup-Crick coefficient for ordination because it yielded the lowest stress value. On the other hand, they primarily used Yule’s Y in their cluster analyses, where:
In a paleoecological and paleogeographical analysis of Late Ordovician cephalopods, Kröger and Ebbestad (2013) used the Raup-Crick and Bray and Curtis coefficients in cluster analysis of assemblages and concluded that the Raup-Crick dissimilarity index gave better-resolved groups.
Balseiro (2016) studied changes in composition and diversity of brachiopods and bivalves in western Argentina during the main Carboniferous glacial event. The author observed few differences among results from several types of ordination and choice of coefficients, including the modified Forbes coefficient of Alroy (2015b) and Bray and Curtis dissimilarity.
Many reviewers of binary coefficients note the controversy that surrounds the question whether mutual absences should be included in a coefficient. Some authors categorically reject coefficients that include d (e.g. Shi 1993). Reasons cited include: mutual absences do not contain information; we can never know the total number of taxa N in a paleogeographic study; we can inflate differences through inappropriate inclusion of taxa or samples; or sampling effort or success is uneven and therefore the appropriate N is unknown. There are counterarguments for each one of these objections and the user is left to decide for his or herself. For instance, knowledge of mutual absences is necessary to evaluate the probability of an observed pattern of occurrences, and therefore it conveys information. While we cannot know N exactly, we have ways to access completeness of sampling, and after all, any statistic is based on samples and N is no exception.
In contrast to the other objections to the use of mutual absences, uneven sampling among locations appears to be a real problem and the effect on even probabilistic measures of association is not well understood. Simpson’s coefficient and modified Forbes coefficient of Alroy (2015a, b) attempt to correct for this problem. Neither coefficient conveys any probabilistic information. This is the price one pays when sampling is less than optimal. To draw strong conclusions sampling methods are all-important.
9 Summary
-
1.
A review of the literature in recent years shows that binary coefficients are still very much used, even given the advantages of abundance information.
-
2.
Choice of coefficient remains a confusing and sometimes empirical process, often leading practitioners to examine results from several coefficients.
-
3.
If a large contrast exists in abundance of taxa or length of faunal lists, one should use care in using similarity coefficients. Comparing dendrograms or ordinations obtained using more than one coefficient could help the practitioner partition out the least informative occurrences.
-
4.
The problem of uneven sampling has not been fully addressed in the literature.
-
5.
There continues to be interest—perhaps growing—in evaluating occurrence data in a probabilistic context.
-
6.
In addition to theoretical considerations, some authors have found empirically that coefficients of association and the related Raup-Crick coefficient work well in clustering and ordination.
In conclusion, it remains a reasonable strategy to compare dendrograms and ordinations calculated with several coefficients. Care should be taken to select coefficients with different performance characteristics. Finally, the practitioner should understand the purpose of each coefficient.
References
Alroy J (2015a) A new twist on a very old binary similarity coefficient. Ecology 96:575–586
Alroy J (2015b) A simple way to improve multivariate analyses of paleoecological data sets. Paleobiology 41:377–386
Balseiro D (2016) Compositional turnover and ecological changes related to the waxing and waning of glaciers during the late Paleozoic ice age in ice-proximal regions (Pennsylvanian, western Argentina). Paleobiology 42:335–357
Brayard A, Escarguel G, Bucher H (2007) The biogeography of early Triassic ammonoid faunas: clusters, gradients, and networks. Geobios 40:749–765
Choi S-S, Cha S-H, Tappert, CC (2009) Correlation analysis of binary similarity and distance measures on different binary database types. In: Proceedings of the international conference on artificial intelligence and pattern recognition, Orlando, Florida, USA, July 2009
Choi S-S, Cha S-H, Tappert CC (2010) A survey of binary similarity and distance measures. J Syst Cybern Inf 8:43–48
Gower JC (1971) A general coefficient of similarity and some of its properties. Biometrics 27:857–871
Gower JC, Legendre P (1986) Metric and Euclidean properties of dissimilarity coefficients. J Classif 3:5–48
Halliday TJD, Prasad GVR, Goswani A (2017) Faunal similarity in Madagascan and south Indian Late Cretaceous vertebrate faunas. Palaeogeogr Palaeoclimatol Palaeoecol 468:70–75
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:9 pp
Hohn ME (1975) Palecology and biostratigraphy of the Portland Point and Kashong Members, Hamilton Group (Middle Devonian) of New York State. Unpublished Master’s Thesis, Indiana University, Bloomington, Indiana, USA, 69 pp
Hohn ME (1976) Binary coefficient: a theoretical and empirical study. Math Geol 8:137–150
Huang B, Rong J, Cocks LRM (2012) Global palaeobiogeographical patterns in brachiopods from survival to recovery after the end-Ordovician mass extinction. Palaeogeogr Palaeoclimatol Palaeoecol 317–318:196–205
Hubálek Z (1982) Coefficients of association and similarity, based on binary (presence-absence) data: an evaluation. Biol Rev 57:669–689
Jackson DA, Somers KM, Harvey HH (1989) Similarity coefficients: measures of co-occurrence and association or simply measures of occurrence? Am Nat 133:486–453
Kröger B, Ebbestad JOR (2013) Palaeoecology and paleogeography of Late Ordovician (Katian-Hirnantian) cephalopods of the Boda Limestone, Siljan district, Sweden. Lethaia 47:15–30
Moore RC et al (1953–2015) Treatise on invertebrate paleontology, parts A through W. Boulder, Colorado: Geological Society of America; and Lawrence, Kansas: University of Kansas Press
Raup DM, Crick RE (1979) Measurement of faunal similarity in paleontology. J Paleontol 53:1213–1227
Schmachtenberg WF (2008) Resolution and limitations of faunal similarity indices of biogeographic data for testing predicted paleogeographic reconstructions and estimating intercontinental distances: a test case of modern and Cretaceous bivalves. Palaeogeogr Palaeoclimatol Palaeoecol 265:255–261
Shi GR (1993) Multivariate data analysis in palaeoecology and palaeobiogeography—a review. Palaeogeogr Palaeoclimatol Palaeoecol 105:199–234
Winrow P, Sutton MD (2014) Lingulate brachiopods and the early Paleozoic history of the Iapetus Ocean. Lethaia 47:456–468
Zhang B, Srihari SN (2003) Properties of binary vector dissimilarity measures. In: Proceedings of the JCIS international conference on computer vision, graphics and image processing, Cary, North Carolina, USA, 26–30 Sept 2003
Acknowledgements
Gordon Baird helped extensively in the early stages of this project by sharing his knowledge of outcrop locations and updated correlations of the Hamilton Group. Thomas Kammer made valuable suggestions to an early draft of this paper.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
<SimplePara><Emphasis Type="Bold">Open Access</Emphasis> This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.</SimplePara> <SimplePara>The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.</SimplePara>
Copyright information
© 2018 The Author(s)
About this chapter
Cite this chapter
Hohn, M.E. (2018). Binary Coefficients Redux. In: Daya Sagar, B., Cheng, Q., Agterberg, F. (eds) Handbook of Mathematical Geosciences. Springer, Cham. https://doi.org/10.1007/978-3-319-78999-6_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-78999-6_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-78998-9
Online ISBN: 978-3-319-78999-6
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)