Abstract
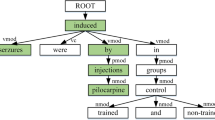
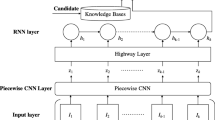
Understanding chemical-disease relations (CDR) from biomedical literature is important for biomedical research and chemical discovery. This paper uses a k-max pooling convolutional neural network (CNN) to exploit word sequences and dependency structures for CDR extraction. Furthermore, an effective weighted context method is proposed to capture semantic information of word sequences. Our system extracts both intra- and inter-sentence level chemical-disease relations, which are merged as the final CDR. Experiments on the BioCreative V CDR dataset show that both word sequences and dependency structures are effective for CDR extraction, and their integration could further improve the extraction performance.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Dogan, R.I., Murray, G.C., Névéol, A., Lu, Z.Y.: Understanding PubMed user search behavior through log analysis. Database (2009), doi:10.1093/database/bap018
Wei, C.H., Peng, Y.F., Leaman, Ret al.: Overview of the biocreative v chemical disease relation (CDR) task. In: The Fifth BioCreative Challenge Evaluation Workshop, pp. 154–166 (2015)
Lowe, D.M., O’Boyle, N.M., Sayle, R.A.: Efficient chemical-disease identification and relationship extraction using Wikipedia to improve recall. Database (2016), doi:10.1093/database/baw039
Xu, J., Wu, Y.H., Zhang, Y.Y., Wang, J.Q., Lee, H., Xu, H.: CD-REST: a system for extracting chemical-induced disease relation in literature. Database (2016), doi:10.1093/database/baw036
Gu, J.H., Qian, L.H and Zhou, G.D.: Chemical-induced disease relation extraction with various linguistic features. Database (2016), doi:10.1093/database/baw042
Pons, E., Becker, B.F.H., Akhondi, S.A., Afzal, Z., van Mulligen, E.M., Kors, J.A.: Extraction of chemical-induced diseases using prior knowledge and textual information. Database (2016), doi:10.1093/database/baw046
Zhou, H.W., Deng, H.J., Chen, L., Yang, Y.L., Jia, C., Huang, D.G.: Exploiting syntactic and semantics information for chemical-disease relation extraction. Database (2016), doi:10.1093/database/baw048
Gers, F.A., Schmidhuber, J.: Recurrent nets that time and count. In: Neural Networks: Como, vol. 3, pp. 189–194 (2000)
Nguyen, T.H., Grishman, R.: Relation extraction: perspective from convolutional neural networks. In: The NAACL Workshop on Vector Space Modeling for NLP, pp. 39–48 (2015)
Kalchbrenner, N., Grefenstette, R., Blunsom, P.: A convolutional neural network for modelling sentences. In: Proceeding of ACL, pp. 655–665 (2014)
Vu, N.T., Adel, H., Gupta, P., Schütze, H.: Combining recurrent and convolutional neural networks for relation classification. In: Proceedings of NAACL-HLT, pp. 534–539 (2016)
Zeng, D.J., Liu, K., Chen, Y.B., Zhao, J.: Distant supervision for relation extraction via piecewise convolutional neural networks. In: Proceedings of EMNLP, pp. 1753–1762 (2015)
Coletti, M.H., Bleich, H.L.: Medical subject headings used to search the biomedical literature. J. Am. Med. Inform. Assoc. 8, 317–323 (2011)
Mikolov, T., Sutskever, I., Chen, K., Corrado, G.S., Dean, J.: Distributed representations of words and phrases and their compositionality. In: Proceedings of NIPS, pp. 3111–3119 (2013)
Xie, R.B., Liu, Z.Y., Sun, M.S.: Representation learning of knowledge graphs with hierarchical types. In: Proceedings of AAAI, pp. 2965–2971 (2016)
Acknowledgements
This research is supported by Natural Science Foundation of China (No. 61272375).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Zhou, H., Yang, Y., Liu, Z., Liu, Z., Men, Y. (2017). Integrating Word Sequences and Dependency Structures for Chemical-Disease Relation Extraction. In: Sun, M., Wang, X., Chang, B., Xiong, D. (eds) Chinese Computational Linguistics and Natural Language Processing Based on Naturally Annotated Big Data. NLP-NABD CCL 2017 2017. Lecture Notes in Computer Science(), vol 10565. Springer, Cham. https://doi.org/10.1007/978-3-319-69005-6_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-69005-6_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-69004-9
Online ISBN: 978-3-319-69005-6
eBook Packages: Computer ScienceComputer Science (R0)