Abstract
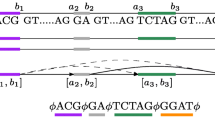
Graphs are the most suited data structure to summarize the transcript isoforms produced by a gene. Such graphs may be modeled by the notion of hypertext, that is a graph where nodes are texts representing the exons of the gene and edges connect consecutive exons of a transcript. Mapping reads obtained by deep transcriptome sequencing to such graphs is crucial to compare reads with an annotation of transcript isoforms and to infer novel events due to alternative splicing at the exonic level.
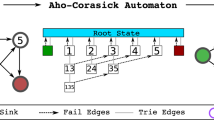
In this paper, we propose an algorithm based on Maximal Exact Matches that efficiently solves the approximate pattern matching of a pattern P to a hypertext H. We implement it into Splicing Graph ALigner (SGAL), a tool that performs an accurate mapping of RNA-seq reads against a graph that is a representation of annotated and potentially new transcripts of a gene. Moreover, we performed an experimental analysis to compare SGAL to a state-of-art tool for spliced alignment (STAR), and to identify novel putative alternative splicing events such as exon skipping directly from mapping reads to the graph. Such analysis shows that our tool is able to perform accurate mapping of reads to exons, with good time and space performance.
The software is freely available at https://github.com/AlgoLab/galig.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Release 29 of ToxoDB annotation of TgondiiGT1.
References
Amir, A., Lewenstein, M., Lewenstein, N.: Pattern matching in hypertext. J. Algorithms 35(1), 82–99 (2000)
Beretta, S., Bonizzoni, P., Della Vedova, G., Pirola, Y., Rizzi, R.: Modeling alternative splicing variants from RNA-seq data with isoform graphs. J. Comput. Biol. 21(1), 16–40 (2014)
Bonizzoni, P., Della Vedova, G., Pirola, Y., Previtali, M., Rizzi, R.: LSG: an external-memory tool to compute string graphs for next-generation sequencing data assembly. J. Comput. Biol. 23(3), 137–149 (2016)
Cormen, T.H., Leiserson, C.E., Rivest, R.L., Stein, C.: Introduction to Algorithms. MIT Press, 2nd edn. (2001)
Dilthey, A., Cox, C., Iqbal, Z., Nelson, M.R., McVean, G.: Improved genome inference in the MHC using a population reference graph. Nat. Genet. 47(6), 682–688 (2015)
Dobin, A., Davis, C.A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M., Gingeras, T.R.: STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1), 15–21 (2013)
Heber, S., Alekseyev, M., Sze, S.H., Tang, H., Pevzner, P.A.: Splicing graphs and EST assembly problem. Bioinformatics 18(suppl. 1), S181–S188 (2002)
Horner, D.S., Pavesi, G., Castrignanò, T., De Meo, P.D., Liuni, S., Sammeth, M., Picardi, E., Pesole, G.: Bioinformatics approaches for genomics and post genomics applications of next-generation sequencing. Briefings Bioinf. 11(2), 181–197 (2010)
Kim, D., Langmead, B., Salzberg, S.L.: HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12(4), 357–360 (2015)
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., Salzberg, S.L.: TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14(4), R36 (2013)
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G.T., Abecasis, G.R., Durbin, R.: The sequence alignment/map format and SAMtools. Bioinformatics 25(16), 2078–2079 (2009)
Manber, U., Wu, S.: Approximate string matching with arbitrary costs for text and hypertext. In: Proceedings of the IAPR International Workshop on Structural and Syntactic Pattern Recognition, pp. 22–33 (1993)
Navarro, G.: Improved approximate pattern matching on hypertext. Theoret. Comput. Sci. 237(1), 455–463 (2000)
Ohlebusch, E., Gog, S., Kügel, A.: Computing matching statistics and maximal exact matches on compressed full-text indexes. In: Chavez, E., Lonardi, S. (eds.) SPIRE 2010. LNCS, vol. 6393, pp. 347–358. Springer, Heidelberg (2010). doi:10.1007/978-3-642-16321-0_36
Rhoads, A., Au, K.F.: PacBio sequencing and its applications. Genomics Proteomics Bioinform. 13(5), 278–289 (2015). sI: Metagenomics of Marine Environments
Sirén, J.: Indexing variation graphs. CoRR abs/1604.06605 (2016)
Thachuk, C.: Indexing hypertext. J. Discrete Algorithms 18, 113–122 (2013)
Trapnell, C., Pachter, L., Salzberg, S.L.: TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25(9), 1105–1111 (2009)
Vyverman, M., De Baets, B., Fack, V., Dawyndt, P.: essaMEM: finding maximal exact matches using enhanced sparse suffix arrays. Bioinformatics 29(6), 802–804 (2013)
Yeoh, L.M., Goodman, C.D., Hall, N.E., van Dooren, G.G., McFadden, G.I., Ralph, S.A.: A serine-arginine-rich (SR) splicing factor modulates alternative splicing of over a thousand genes in Toxoplasma gondii. Nucleic Acids Res. 43(9), 4661–4675 (2015)
Acknowledgments
We thank the anonymous reviewers for their insightful comments.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Beretta, S., Bonizzoni, P., Denti, L., Previtali, M., Rizzi, R. (2017). Mapping RNA-seq Data to a Transcript Graph via Approximate Pattern Matching to a Hypertext. In: Figueiredo, D., Martín-Vide, C., Pratas, D., Vega-Rodríguez, M. (eds) Algorithms for Computational Biology. AlCoB 2017. Lecture Notes in Computer Science(), vol 10252. Springer, Cham. https://doi.org/10.1007/978-3-319-58163-7_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-58163-7_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-58162-0
Online ISBN: 978-3-319-58163-7
eBook Packages: Computer ScienceComputer Science (R0)